Once upon a time, a colleague of mine shared a rather intriguing story. They had proposed to their boss to switch from testing for fecal coliforms to using a substrate enzyme medium that utilizes β-glucuronidase activity. However, their boss shot down the idea with the objection that this medium wouldn’t cover the infamous E. coli O157:H7. So, what’s the real scoop here? Are we overlooking some vital points in our understanding?

The Principle Behind β-Glucuronidase-Based Enzyme Substrate Media
The detection of E. coli using β-glucuronidase-based enzyme substrate media banks on the unique enzyme activity present in these bacteria. β-Glucuronidase is an enzyme that acts on the β-glycosidic bond of D-glucuronic acid glucuronides, hydrolyzing them to release glucuronic acid.
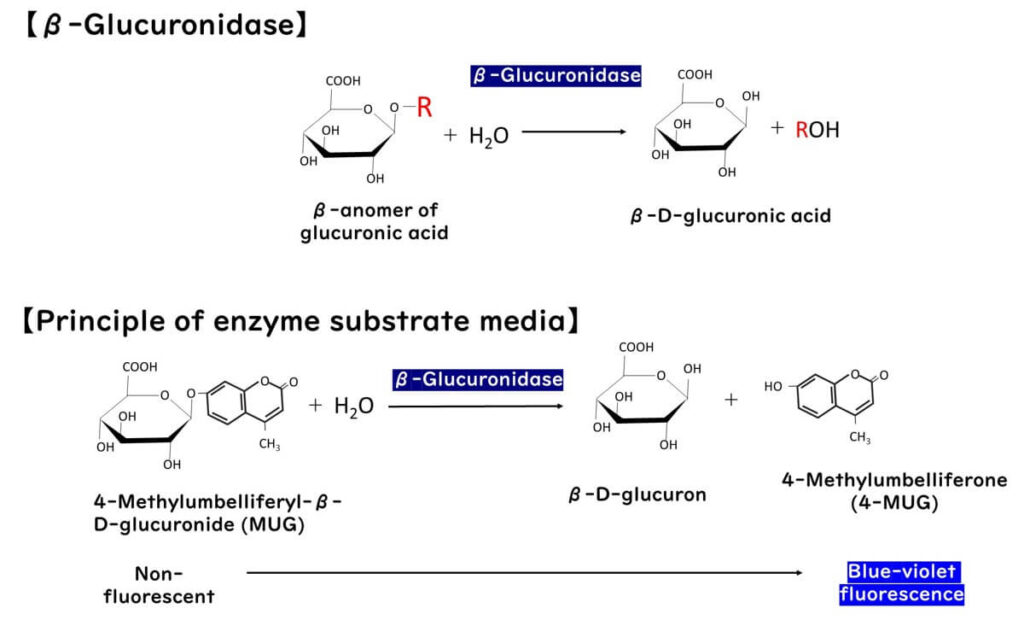
During this process, specific chromogenic substrates decompose in the presence of this enzyme, leading to color changes or fluorescence.
As colorimetric enzyme substrates for β-glucuronidase, compounds like MUG (4-methylumbelliferyl-β-D-glucuronide) and X-Glucuronide (5-bromo-4-chloro-3-indolyl-β-D-glucuronide) are utilized. E. coli cells can absorb these compounds intact, and the intracellular glucuronidase cleaves their glucuronide bonds. The resulting MUG emits fluorescence under long-wave ultraviolet light, while X-Glucuronide turns blue. Thus, the presence and quantity of E. coli can be indirectly detected by visually observing the color change or fluorescence following the β-glucuronidase reaction. Based on this principle, various media have been developed.
It's noteworthy that β-glucuronidase activity is predominantly found in Escherichia coli (E. coli) among Gram-negative bacteria, with 90 to 96% of strains exhibiting this trai(Thompson et al.,1990、Hansen et al.,1984). While other bacteria like Shigella, Yersinia, and some Salmonella strains may also possess this enzymatic activity, it is primarily produced by E. coli, and its presence in other enterobacterial species is rare. For instance, Thompson et al.,(1990) reported that out of 421 non-E. coli bacterial strains isolated from human clinical specimens, only 3 out of 92 Salmonella strains tested positive for MUG, and for Shigella, only 11 out of 14 Shigella sonnei isolates, 1 out of 5 Shigella boydii isolates, and 1 out of 17 Shigella flexneri isolates showed MUG positivity. Additionally, no positive results were found among 12 isolates of Yersinia enterocolitica.
Enzyme Substrate Media Can Detect Non-O157:H7 STEC Strains
As previously discussed, it's established that 90 to 96% of E. coli strains, regardless of their pathogenicity, possess β-glucuronidase activity. However, the serotype O157:H7 clone is a rare exception to this rule.
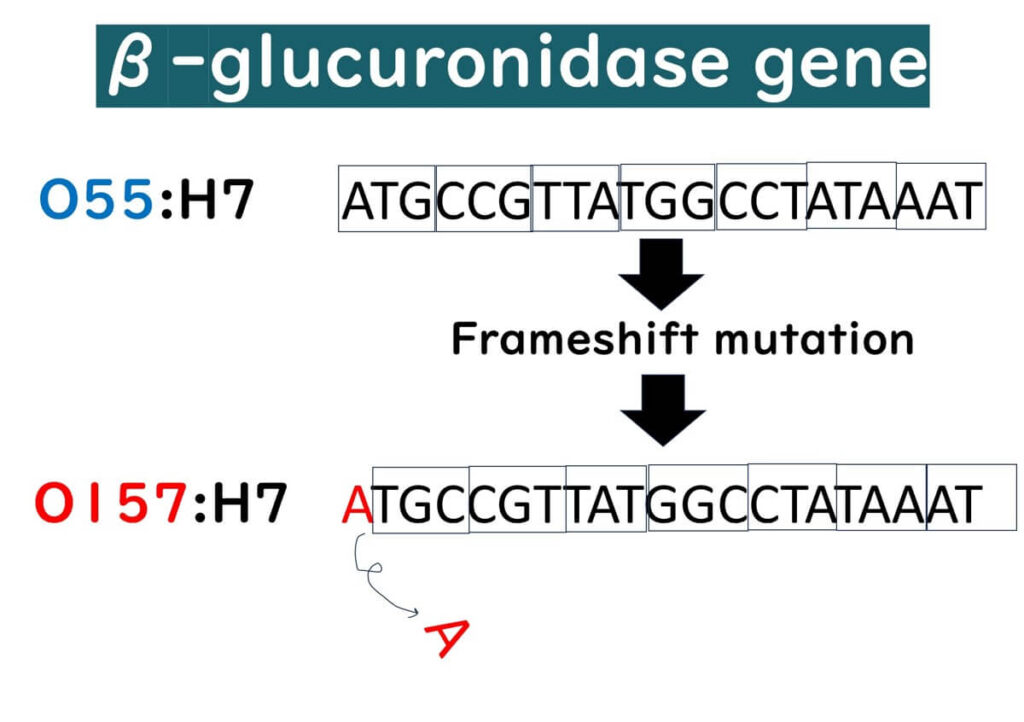
The primary reason most O157:H7 strains lack β-glucuronidase activity is believed to be evolutionary. Originating from O55:H7, which is positive for β-glucuronidase, O157:H7 has progressively lost the ability to ferment sorbitol and β-glucuronidase activity(Wick et al., 2005). A frameshift mutation in the β-glucuronidase gene of O55:H7 is thought to have led to the loss of this enzyme expression. This loss can be seen as a specific evolutionary genetic mutation unique to O157:H7.
Despite this, it's essential to clarify that the absence of β-glucuronidase is generally only characteristic of the enterohemorrhagic E. coli O157:H7. There has been less research concerning other serotypes of enterohemorrhagic E. coli and their β-glucuronidase activity, making it difficult to definitively state whether they lack this activity in the same way as O157:H7. Indeed, other serotypes may have evolved to lose β-glucuronidase activity, but this area remains under-researched.
Several cases where β-glucuronidase activity was positive include:
- In July 2014, an outbreak of Shiga toxin-producing E. coli (STEC) O55:H7 occurred in the UK, affecting 31 people, with 13 (42%) developing hemolytic uremic syndrome. This strain was reported to be β-glucuronidase positive (Schutz et al, 2017).
- A study of 49 strains of STEC O121:H19 related to flour outbreaks in the United States and Canada found that, except for six strains, all showed normal β-glucuronidase activity (Gill et al.,2022).
- An examination of various serotypes of Shiga toxin-producing E. coli strains (O1:H-, O111:H8, O111:H-, O113:H21, O26:K60, O91:H21, O121:H19, O145:H-, and untypable strains) showed that β-glucuronidase reactions varied from positive to negative Krishnan et al.,1987).
- Research on STEC isolates from cattle in Chile, not including serotype O157:H7, found that 96.8% (90 out of 93) were positive for β-glucuronidase activity(Diaz et al., 2021).
However, there are also reports of β-glucuronidase-negative strains among non-O157 serotypes.
- For instance, an STEC O26 isolated from a 4-year-old girl in Yamaguchi Prefecture, Japan, in 2012, was reported as β-glucuronidase-negative(Kameyama et al.,2013).
In summary, while most non-O157 serotypes of enterohemorrhagic E. coli are typically β-glucuronidase-positive, some negative strains do exist (based on current data).
It's crucial to recognize that the limitations of β-glucuronidase-based enzyme substrate media are primarily confined to the serotype O157 and do not extend to other types of enterohemorrhagic E. coli.
Even Fecal Coliform Testing Can Miss Some O157:H7 Strains
Believe it or not, the issue with failing to detect E. coli O157:H7 isn't unique to enzyme substrate media. It's also a known complication in fecal coliform tests conducted at 44.5°C. According to the international standard ISO 16649-1:2018, the cultivation temperature for detecting E. coli is set at 44°C. However, some strains of E. coli O157:H7 struggle to proliferate at this temperature, virtually making them undetectable using this method (ISO 16649-1:201).
In the world of food company self-testing, when there's a proposal to switch from fecal coliform media to enzyme substrate media, there's often hesitance due to the explicit mention in enzyme substrate media manuals that detection of O157:H7 may not be covered. This concern mirrors the situation described earlier where a boss might hesitate to change the testing approach. However, the reality that not all E. coli O157:H7 can be detected even at the fecal coliform test's optimal temperature of 44.5°C should prompt a reconsideration of the criticisms aimed at enzyme substrate media.

Understanding that the fecal coliform testing method at 44.5°C does not guarantee the detection of all E. coli O157:H7 strains should lead us to question the accuracy of viewing enzyme substrate media as inferior. If both methods have their limitations in detecting this particular pathogen, it underscores the need for a more nuanced approach to microbial testing, perhaps incorporating multiple testing strategies to ensure food safety comprehensively.
E. coli Testing as an Indicator Organism Does Not Primarily Aim to Detect O157:H7
A crucial understanding in the realm of microbial testing is that E. coli tests, particularly those using indicator organisms, do not primarily aim to detect O157:H7. This simple fact, often misunderstood by many food company quality control personnel, warrants a fresh discussion about the purpose of testing E. coli.
What exactly is an indicator organism? An indicator organism is used to suggest the possible presence of pathogens that share similar environmental behaviors. This means the bacteria detected by the culture media and the pathogens inferred from this presence are fundamentally different.
Another commonly overlooked point among quality managers is the difficulty of accurately assessing the contamination risk of pathogens like E. coli O157:H7, which are present in small amounts. For instance, if the contamination rate of a pathogen is 1%, analyzing a single sample means there's a 99% chance of overlooking the contamination (false negative). Even analyzing 30 samples results in a 74% chance of a false negative. To avoid missing the contamination with 95% accuracy, it requires analyzing 298 samples. For a more detailed explanation, one might consider reading introductory articles on the purpose and calculation methods of microbial testing in foods.
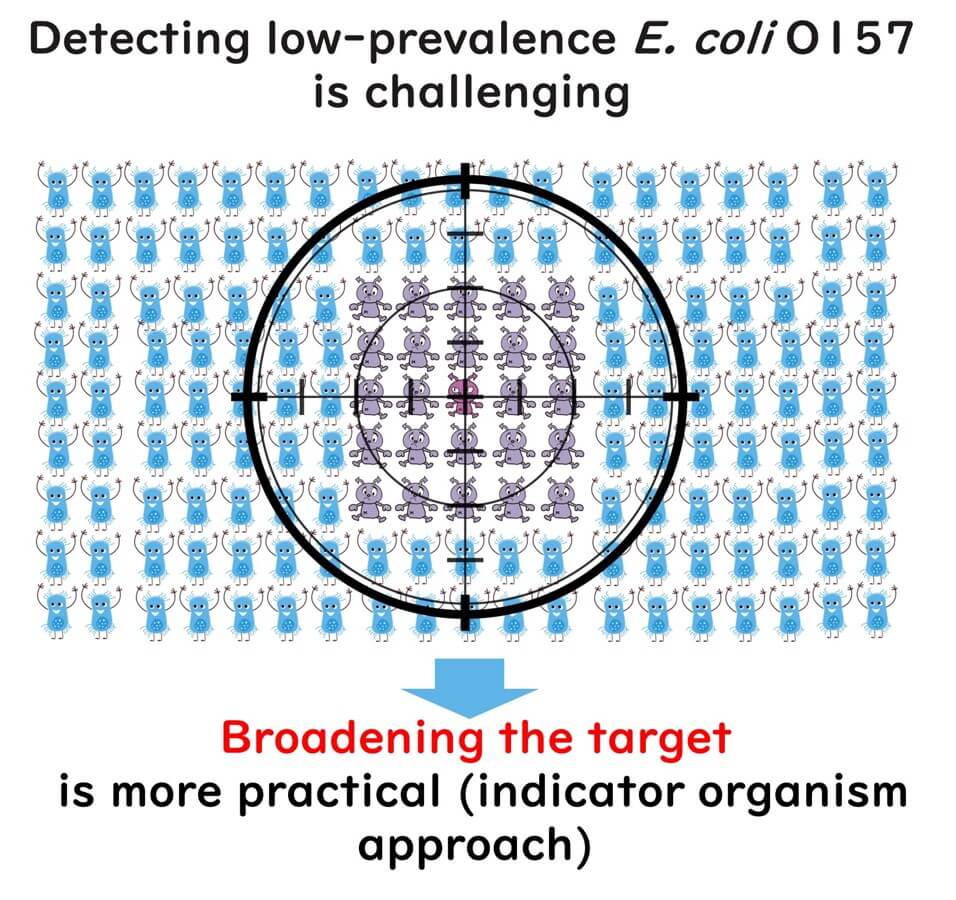
Thus, when the distribution of a target pathogen is sparse, analyzing a few samples hardly serves the intended purpose of testing. Instead, more prevalent bacteria such as E. coli are detected as indicator organisms. Even though the issues of sample numbers and false negatives persist even with indicator organism detection, the higher distribution rate of these bacteria (and consequently, a lower rate of false negatives) justifies their use.
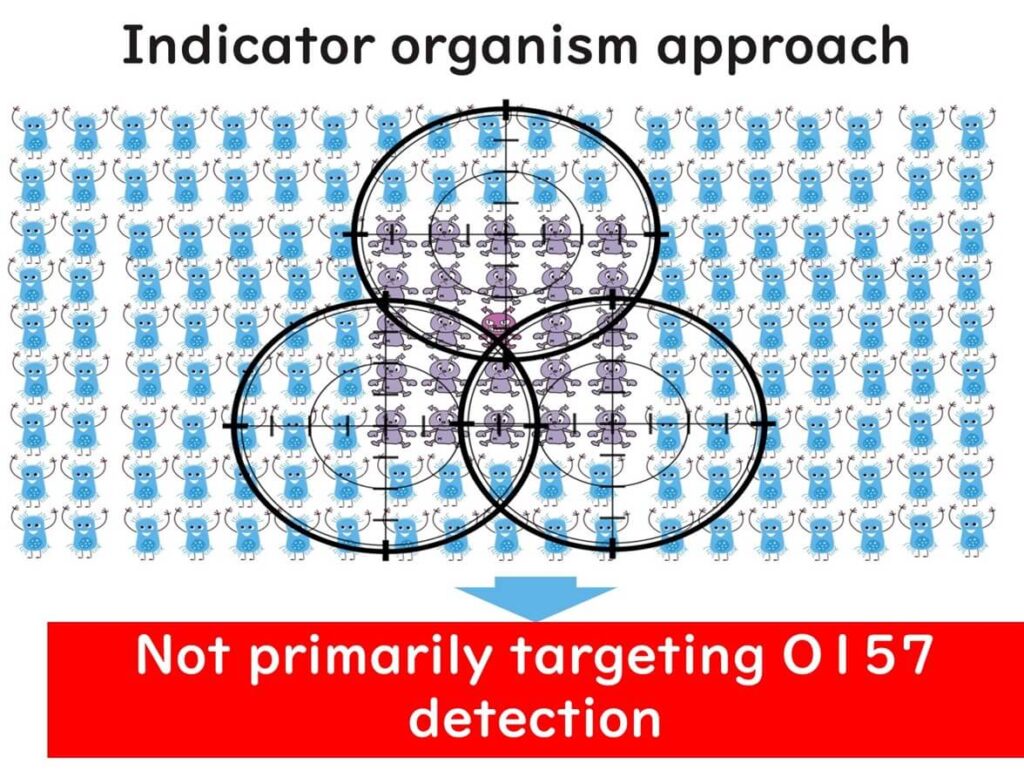
In sum, it's essential to grasp the significance of indicator organism testing. Therefore, even if E. coli O157:H7 is not detected on enzyme substrate media used for E. coli tests, the primary goal of indicator organism testing isn't to detect pathogens on agar media directly. This is a common point of confusion and error among the quality managers I have consulted with.
Summary
This article was written to refresh our understanding of the significance of using enzyme substrate media in E. coli testing as indicator organisms. As mentioned at the outset, enzyme substrate media are not suitable for detecting E. coli O157:H7, which sometimes causes hesitation among quality control personnel. It is hoped that this article will clear up any misconceptions and assist quality managers in making informed decisions about the use of enzyme substrate media. This knowledge empowers them to utilize these tools effectively, enhancing their microbial testing strategies while understanding their limitations and scopes.