Achieving accuracy in PCR food testing is critical for reliable microbial detection. This page explores common pitfalls that can lead to false positives and negatives, such as contamination, PCR inhibitors, and the lack of proper validation. By understanding these challenges and implementing effective precautions, food safety professionals can ensure dependable results and uphold the highest standards in quality control.Factors Inhibiting Gene Amplification (False Negatives)
Introduction: Addressing the Challenges of PCR Reliability
PCR testing has revolutionized microbial detection in food microbiology, offering precision and speed unmatched by traditional methods. However, like any analytical tool, PCR is not without its limitations. False negatives, often caused by inhibitory substances in food samples, can compromise the reliability of results. To ensure accurate detection, it is essential to recognize these challenges and implement effective measures to mitigate them. This section delves into the factors that inhibit gene amplification and explores strategies to overcome these barriers, paving the way for more reliable PCR-based testing.
Factors Inhibiting Gene Amplification (False Negatives)
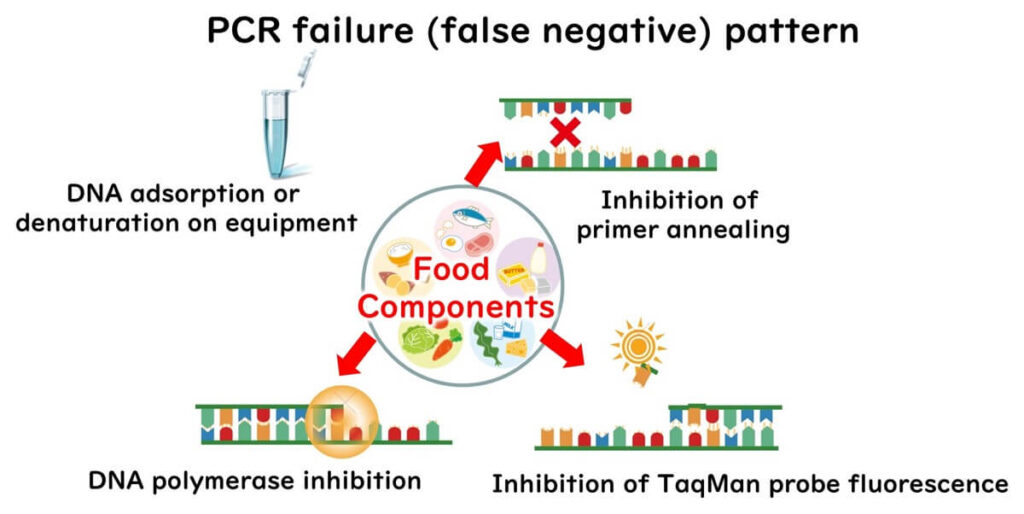
In genetic testing, various components derived from food can inhibit gene amplification during both the extraction and amplification processes. These factors can be broadly categorized as follows:
- Adsorption of Target DNA: During the extraction process, target DNA may adhere to the container walls, leading to its denaturation and loss.
- Inhibition of Primer Binding: Components in the amplification mix may interfere with the primers' ability to bind to the target DNA.
- Inhibition of DNA Polymerase: The enzymatic activity of DNA polymerase may be disrupted by certain inhibitors present in the food matrix.
- Inhibition of Fluorescent Probes: When using fluorescent probes such as TaqMan, substances in the sample may suppress fluorescence emission, affecting the detection process.
In food microbiology, these PCR inhibitors are a critical consideration when attempting to detect specific microorganisms in food samples. If PCR results are negative, it can be difficult to determine whether the target microorganism is genuinely absent or if inhibitors in the food have prevented amplification.
To mitigate this issue, an internal positive control (IPC) with a known gene sequence is included in the PCR reaction. Amplification of the IPC confirms that PCR inhibitors in the food did not interfere with the reaction.
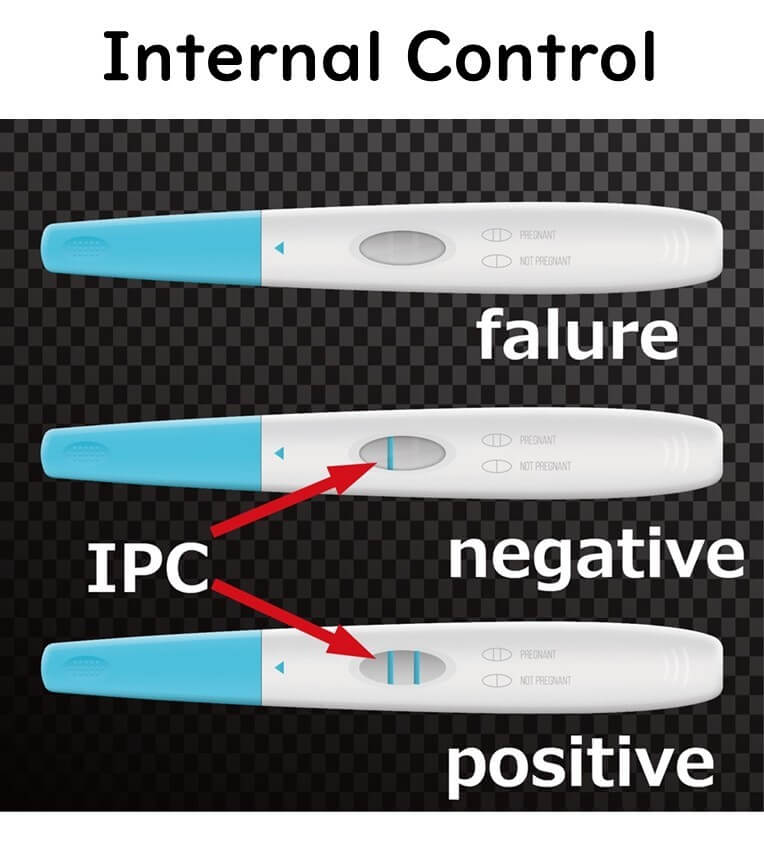
There are two main types of internal controls:
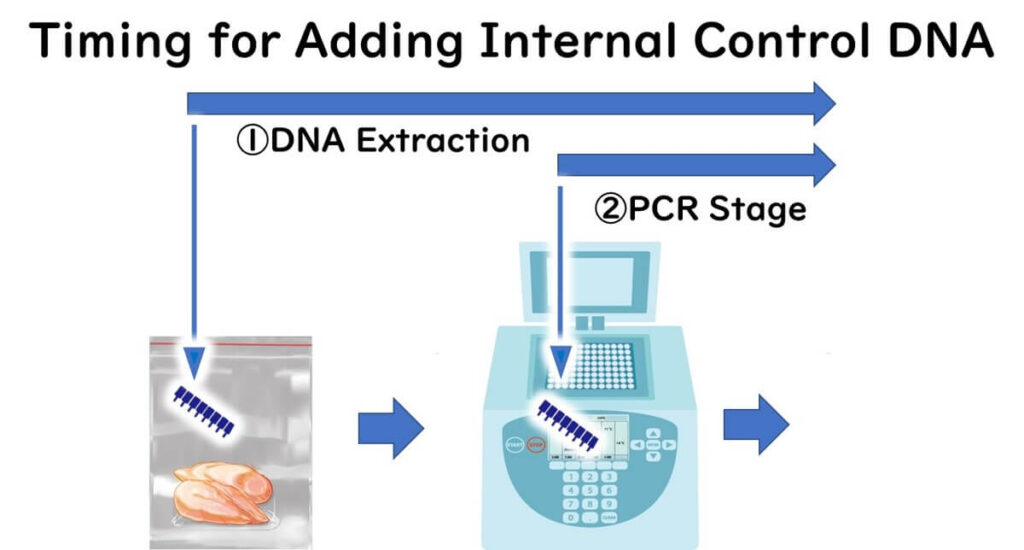
- Controls added during the food extraction stage: These verify the entire process, from DNA extraction to amplification.
- Controls added during the gene amplification stage only: These ensure the amplification process itself is not inhibited.
While adding gene fragments is more common in practice, a more robust approach involves inoculating a standard bacterial strain directly into the food sample. This method allows for comprehensive verification starting from the extraction stage, ensuring that the process is effective even in complex food matrices.
Attention to Gene Contamination (False Positives)
False positives in PCR testing can significantly compromise the reliability of results. These occur when genes not originally present in the sample are mistakenly introduced during the testing process. The primary causes of false positives can be categorized as follows:

- Contamination Between Different Samples: Genes from one sample may contaminate another during handling.
- Contamination from Amplified Samples: Amplified DNA from previous tests can contaminate new samples.

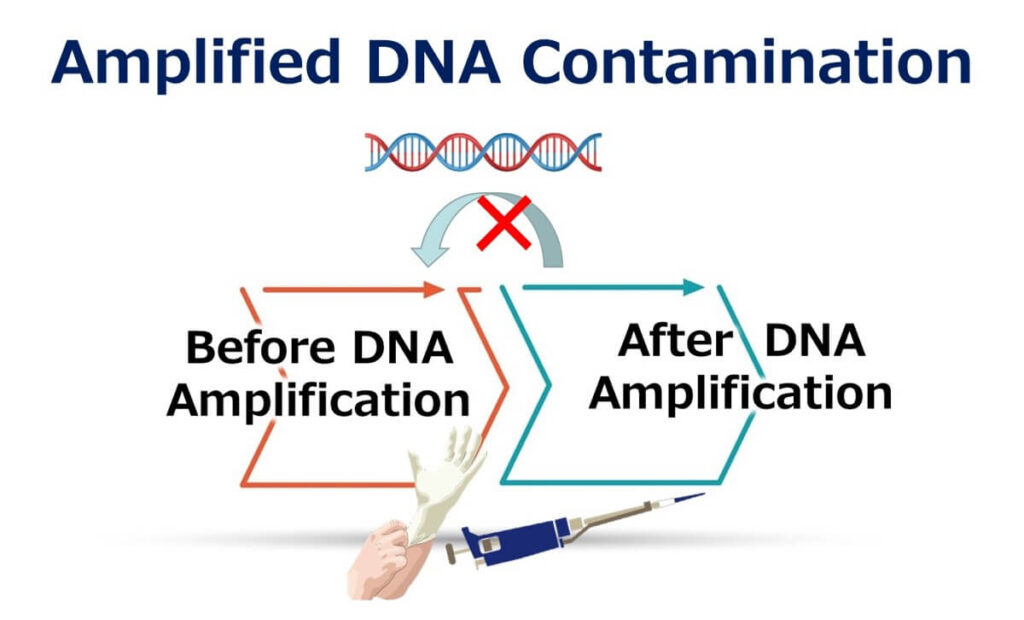
To mitigate these risks, stringent laboratory practices are essential:
Proper Handling Techniques: Pipetting should be done carefully to prevent the formation and spread of aerosols.
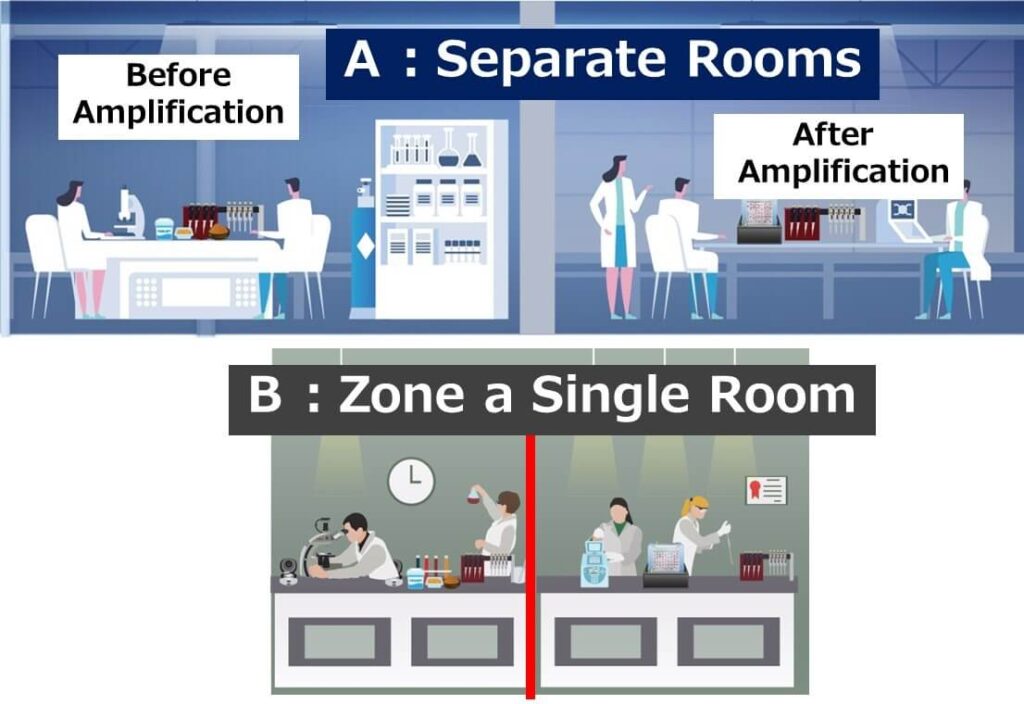
Laboratory Zoning: Ideally, separate the areas for sample preparation and post-amplification analysis. If this is not feasible, establish clear zones within the same laboratory.
Dedicated Equipment: Avoid sharing pipettes, glassware, gloves, and lab coats between zones to minimize cross-contamination.
Workplace Cleanliness: Regularly clean workspaces with disinfectants like sodium hypochlorite before beginning experiments.
Additionally, regular training for laboratory personnel ensures these precautions are consistently implemented.
For robust analysis, include a No Template Control (negative control) in every experimental setup. This control does not contain the target gene and helps identify potential contamination. If frequent positive reactions occur in these controls, it signals a fundamental issue with lab management that needs immediate attention.
Detecting Microorganisms from Food is More Challenging than Clinical Detection
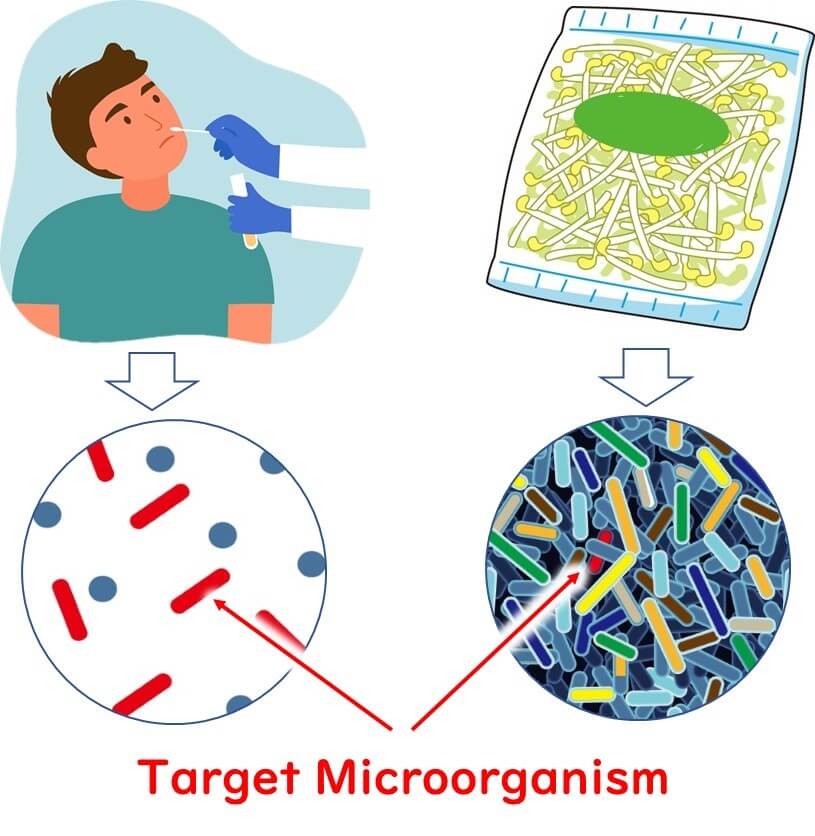
Detecting pathogens using PCR in food samples presents unique challenges compared to clinical samples. These differences arise primarily due to the complexity and diversity of microorganisms present in food matrices.
In clinical samples, such as blood, nasal, or throat swabs, the variety of coexisting microorganisms is relatively limited. This simplifies the detection process. Conversely, food samples often contain a wide array of microorganisms, including both harmless and potentially harmful species. Detecting a specific target microorganism, which may exist as a single cell (1 cfu) in 25 grams of food, becomes significantly more difficult under these conditions.
To address these challenges, precise primer design and careful pre-PCR steps are essential. These include:
- Employing selective enrichment cultures to increase the target bacteria population relative to other microorganisms.
- Optimizing sample preparation to minimize inhibitors and maximize the likelihood of detecting the target pathogen.
By implementing these measures, food safety professionals can improve the reliability of PCR-based microbial detection in food samples, overcoming the inherent complexities of food matrices.
PCR Detection Without Validation is Unreliable
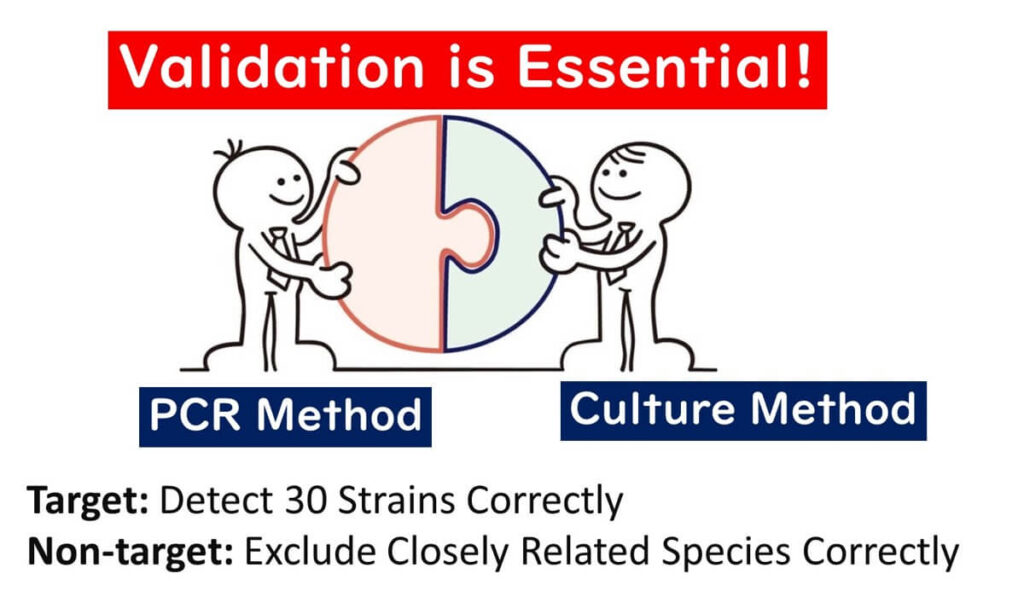
As discussed in this article, PCR testing in food microbiology presents inherent risks. To mitigate these risks and ensure accuracy, a robust validation process is indispensable. Validation involves extensive testing with multiple strains to confirm both the sensitivity and specificity of the method.
For example, international standards such as ISO 16140:2003 and AOAC guidelines set clear validation criteria for target and non-target strains:
- Target species strains: At least 30 strains
- Non-target species strains: At least 30 strains
For certain bacteria, these numbers may increase to 50 strains per category. Validation ensures that the method reliably identifies the target species while minimizing false positives or negatives.
In genetic testing, rigorous validation is non-negotiable. Methods that lack comprehensive validation should be considered unreliable and unsuitable for critical applications such as food safety.
Challenges in Published Research As an editor for international journals, I have reviewed numerous papers proposing new PCR primers for microorganism detection. A recurring issue is insufficient validation—many studies claim specificity based on tests with fewer than ten strains. Such limited validation is inadequate for food microbiology, where reliability is paramount.
Key Takeaway When adopting PCR methods for food microbiology testing, always ensure that they meet established validation standards, such as those outlined by ISO or AOAC. This step is critical for maintaining the credibility and reliability of your testing processes.

PCR Testing Work Schedule
To conclude, here is a practical work schedule for performing microbiological tests on food samples using PCR.
Challenges in Routine Inspections
In routine inspections at food manufacturing and distribution facilities, the primary focus is often on detecting whether there is as little as one cell of a foodborne pathogen per 25 grams of food. This poses a unique challenge because the quantity of the target gene in such samples is extremely low. Even with PCR, an enrichment culture step is essential to amplify the pathogen to a detectable level.
Unlike traditional culture methods, PCR requires less time for enrichment, typically only a few hours. This efficiency makes it possible to complete the entire analysis within a single workday. A recommended schedule is outlined below:
Day 1: Evening
- Mix the food sample (25g) with 225ml of enrichment medium.
- Homogenize the mixture using a stomacher.
- Incubate the mixture at 37°C overnight.
Day 2: Morning (9 AM)
- Start the analysis promptly.
- Perform DNA extraction, PCR amplification, and result interpretation.
- Aim to complete all steps by the end of the morning.
Efficiency for HACCP Implementation
By following this schedule, food safety professionals can determine the presence of foodborne pathogens in food samples daily. This rapid testing approach is invaluable in food manufacturing environments that implement HACCP (Hazard Analysis Critical Control Points). It enables timely microbiological quality assessments of raw materials and intermediate products, supporting proactive decision-making.