To understand DNA sequencing in food microbiology, it is essential to grasp how different types of DNA sequencers function. This article provides a clear overview of key sequencing methods, including the Sanger method (electrophoresis, capillary sequencer), next-generation sequencing (NGS) (Illumina, Ion Torrent), and third-generation sequencing (TGS) (nanopore sequencing).Designed for beginners in food microbiology, this guide explains these technologies in a visual and accessible way, with video demonstrations for clarity. We will also explore the practical applications of DNA sequencing in food safety and microbiological testing.
What is the Sanger Method? History and Principles
DNA sequencing was pioneered by Dr. Frederick Sanger in the UK. The Sanger sequencing method, named after him, earned him the Nobel Prize in Chemistry in 1980. Interestingly, before developing DNA sequencing, Dr. Sanger had already created a method for determining protein sequences, which earned him his first Nobel Prize in 1958. As a result, he became one of the few scientists to receive two Nobel Prizes for distinct scientific achievements.
How the Sanger Method Uses Electrophoresis
The Sanger sequencing method determines DNA sequences using Polymerase Chain Reaction (PCR). While Dr. Sanger's original method predates PCR, explaining the modern capillary sequencer using PCR terminology makes it easier to understand.
Unlike standard PCR, which uses two primers, Sanger sequencing employs only one primer, leading to a single-direction extension reaction. However, simply extending the DNA does not provide sequencing information—the key lies in how the reaction is controlled.
Key Principle of the Sanger Method
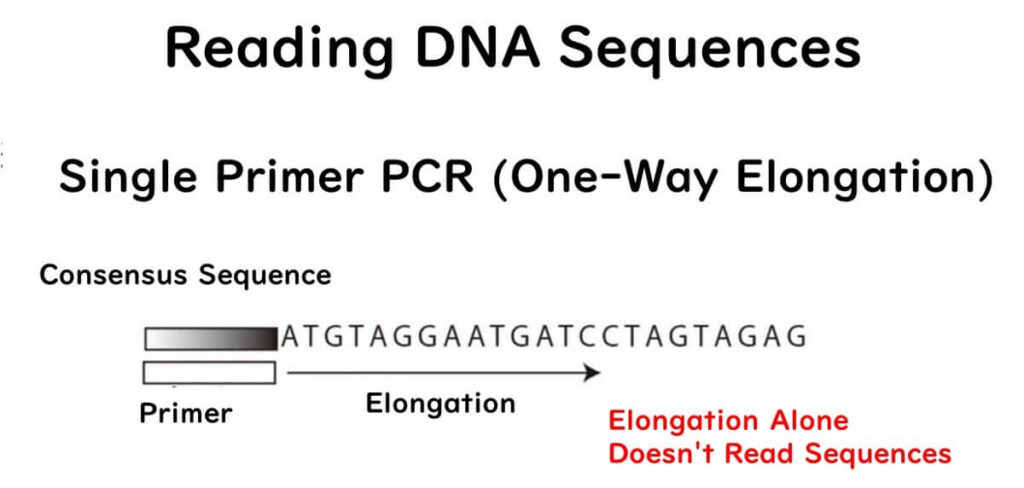
During DNA synthesis, four types of nucleotides (A, T, C, G) are required. These nucleotides are the building blocks of DNA, consisting of a base, sugar, and phosphate group.
Here’s where Dr. Sanger's key innovation comes in:
- By introducing modified nucleotides (lacking a 3’ OH group), the DNA polymerase cannot extend the DNA further when it incorporates them.
- This results in DNA fragments of various lengths, stopping at different positions where these defective nucleotides are incorporated.
- To ensure proper sequencing, a mixture of mostly normal nucleotides and a small proportion of modified nucleotides is used.
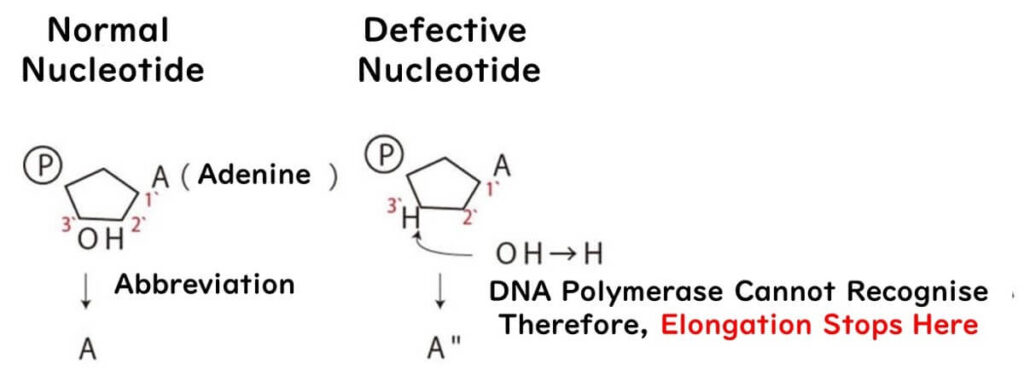
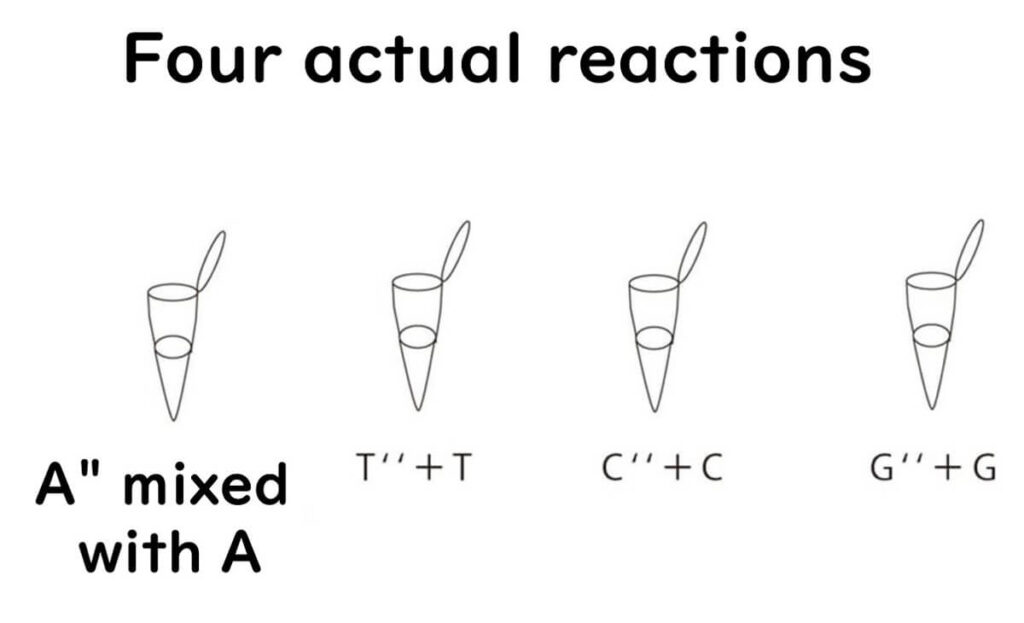
For example, when defective A nucleotides are introduced:
- If the concentration of defective A is low, longer DNA fragments are produced.
- If the concentration is high, shorter fragments dominate.
- A balanced mix produces a full spectrum of fragment lengths.
Electrophoresis for DNA Separation
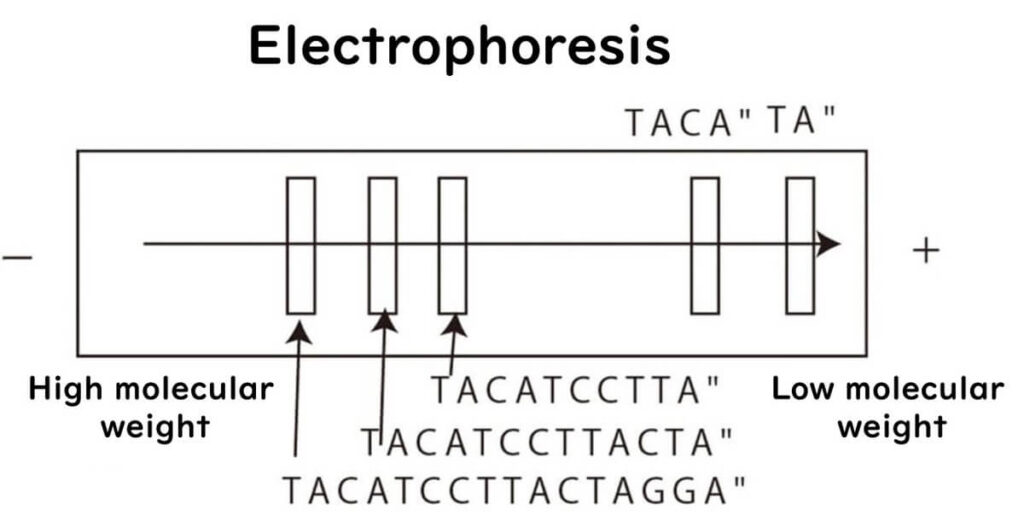
Once the DNA fragments are generated, they are separated using electrophoresis:
- Smaller fragments travel faster and farther through the gel.
- Larger fragments move slower and stay near the starting point.
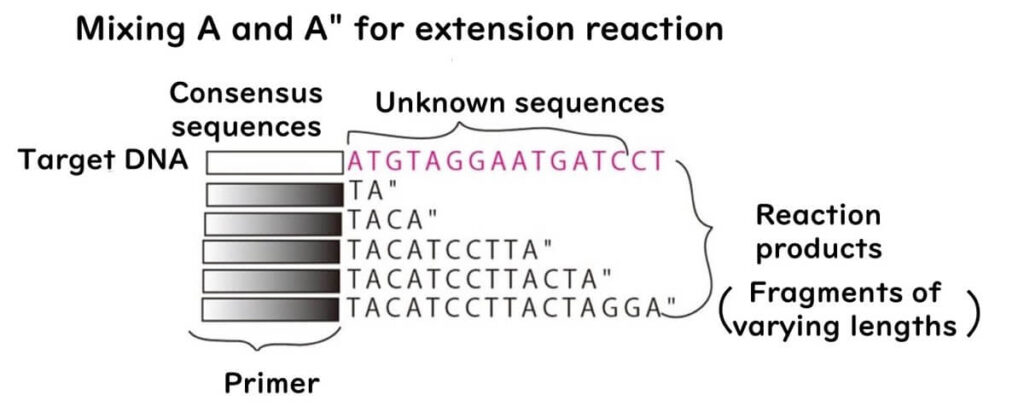
This process is repeated individually for each nucleotide (A, T, C, G) using four separate reaction mixtures.
During electrophoresis, DNA fragments are run on four separate lanes corresponding to the four nucleotides.
- The smallest fragments appear closest to the positive electrode.
- The bands never overlap across lanes, allowing for accurate sequence determination.
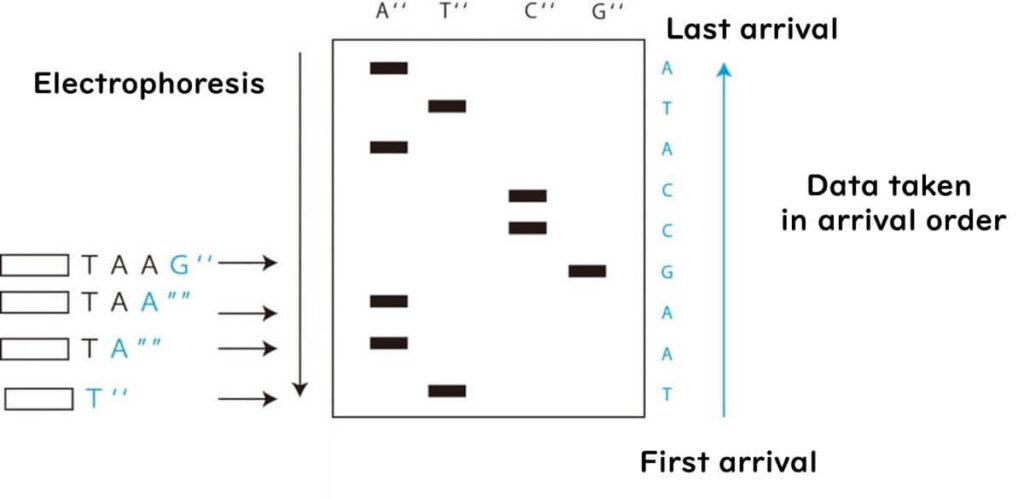
Decoding the DNA Sequence
By reading the bands from the smallest to the largest and identifying the corresponding lane, the complementary sequence of the original DNA strand is determined.
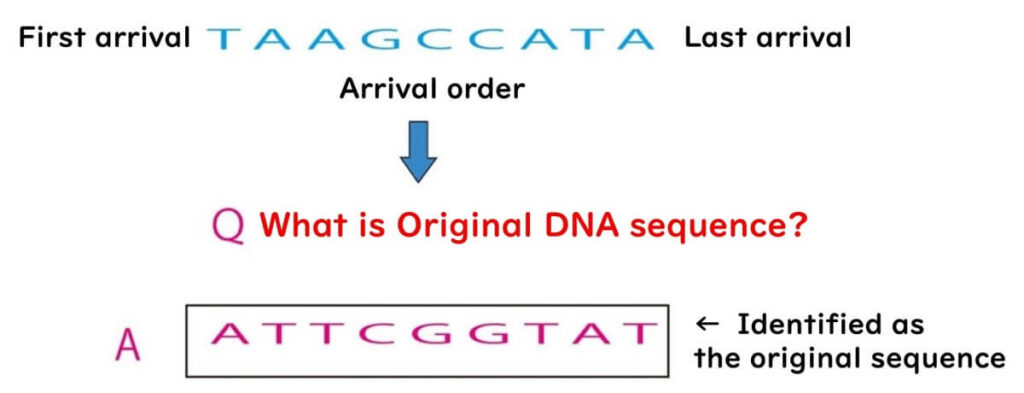
Reversing this complementary sequence provides the original DNA sequence.
This classical Sanger sequencing method forms the foundation for modern DNA sequencing technologies.
The Role of Flat Gel Electrophoresis Sequencers in Food Microbiology
In early DNA sequencing, electrophoresis was stopped at intervals to analyze DNA fragments one step at a time. However, in the early 1990s, a new method emerged that used fluorescently labeled nucleotides (A, T, C, G). Each nucleotide was tagged with a unique fluorescent dye, allowing continuous electrophoresis without needing to pause the process.
This innovation led to the development of flat gel electrophoresis fluorescent dye DNA sequencers, which became widely used during the 1990s. However, by the late 1990s, capillary sequencers replaced this technology, making flat gel electrophoresis sequencers obsolete.
Flat Gel Electrophoresis in Food Microbiology Testing
Although flat gel electrophoresis sequencers were short-lived in food microbiology testing, they played a valuable role in education. Their visual clarity made them ideal for demonstrating the Sanger sequencing method to students.
For nearly a decade after capillary sequencers were introduced, I used flat gel electrophoresis fluorescent sequencers for third-year food microbiology laboratory experiments at university. Their simplicity and visual appeal made them effective teaching tools. However, by 2012, we had completely transitioned to capillary sequencers for educational purposes, marking the end of flat gel electrophoresis sequencing in my laboratory.
How Capillary Sequencers Revolutionized DNA Sequencing
By the mid-1990s, capillary sequencers were introduced, marking a major advancement in DNA sequencing technology. These sequencers were sold by Applied Biosystems (ABI) but were based on core technology developed by Hitachi.
In capillary sequencing, the four DNA bases (A, T, C, G) are separated within a thin capillary tube. Each base is fluorescently labeled with a unique dye, allowing detection of fragment sizes and sequence determination. As DNA fragments move through the capillary, they emit distinct fluorescent signals, which are read by the sequencer to determine the DNA sequence.
For an overview of capillary sequencer principles, please watch the video below.
Applications of Capillary Sequencers in Food Microbiology
Since the late 1990s, capillary sequencers have been the gold standard for genetic testing in food microbiology. While next-generation sequencers (NGS)—discussed later—are increasingly used for whole-genome sequencing (WGS), 16S rDNA metagenomics, and molecular epidemiology, capillary sequencers remain essential for targeted gene sequencing.
Key Applications in Food Microbiology:
✅ Microbial Identification – Capillary sequencers are widely used to identify microbial strains in food safety testing, particularly through 16S rDNA sequencing.
✅ Molecular Epidemiology – In Japan, capillary sequencing has played a key role in tracking outbreaks of enterohemorrhagic Escherichia coli (EHEC) since 2018.
✅ Foodborne Pathogen Source Tracing – Capillary sequencing is used to identify infection sources of serotypes O157, O26, and O111 using the MLVA (Multiple-Locus Variable Number Tandem Repeat Analysis) method.
Although NGS technologies are advancing rapidly, capillary sequencers remain highly relevant in food microbiology testing, particularly for routine pathogen identification and strain differentiation.
How Next-Generation Sequencers (NGS) Transformed DNA Sequencing
Since their emergence in the late 1990s, capillary sequencers have been the gold standard in DNA sequencing. However, in 2007, a groundbreaking technology called Next-Generation Sequencing (NGS) was introduced, fundamentally changing the field.
NGS offers unprecedented speed and cost efficiency, making it vastly superior to the Sanger method. Unlike traditional sequencing, which analyzes DNA one fragment at a time, NGS processes millions of DNA fragments simultaneously, revolutionizing genetic analysis.
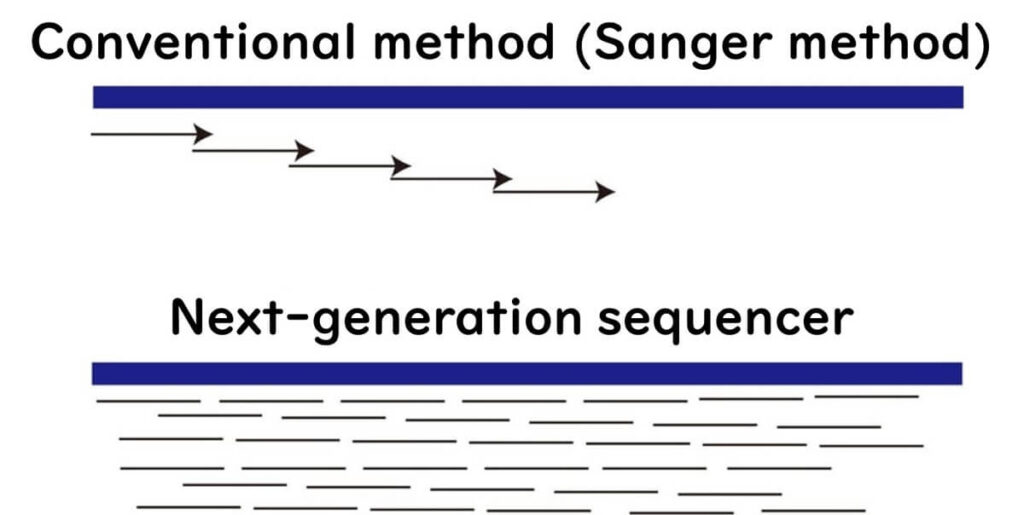
How NGS Works: A Simple Analogy
Imagine trying to read a book with tens of thousands of pages using the Sanger method—it would take years to complete.
In contrast, NGS works like giving each page to thousands of readers, allowing the book to be fully read in a single day.
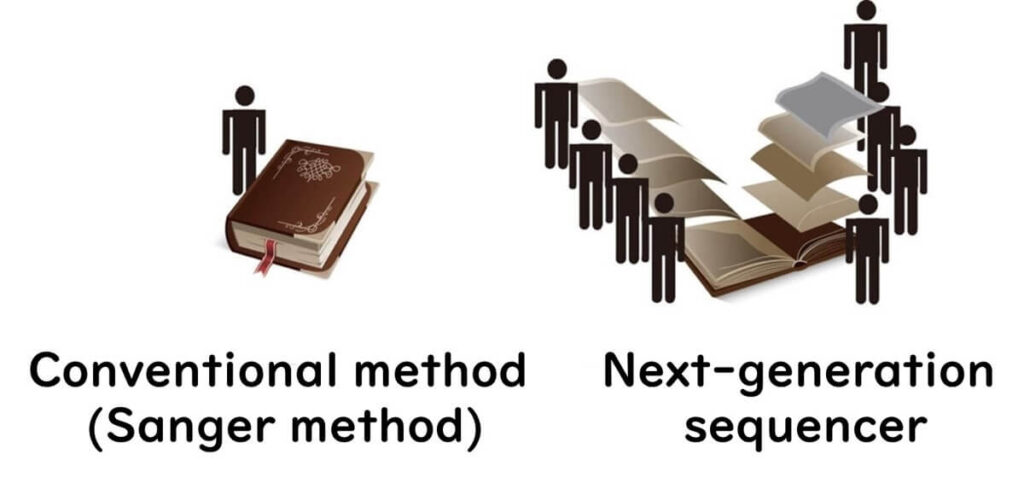
This advancement allows NGS to determine DNA sequences thousands to tens of thousands of times faster than previous methods.
Key Steps in Next-Generation Sequencing:
1️⃣ DNA Fragmentation – The genome of a microorganism is broken down into short DNA fragments (~300 bases long).
2️⃣ Parallel Sequencing – Millions of these fragments are simultaneously sequenced on a microarray platform.
3️⃣ Data Assembly – Advanced software reconstructs the entire genome from these short sequences.
Below, we explore two major NGS technologies:
✅ Illumina Sequencing
✅ Ion Torrent Sequencing
Illumina Sequencers
Illumina sequencing is one of the most widely used Next-Generation Sequencing (NGS) technologies. It enables high-throughput DNA sequencing with exceptional accuracy, making it a gold standard in food microbiology and genetic research.
Key Steps in Illumina Sequencing:
1️⃣ DNA Fragmentation – The genome of the target microorganism is fragmented into short pieces (~200 base pairs) using restriction enzymes.
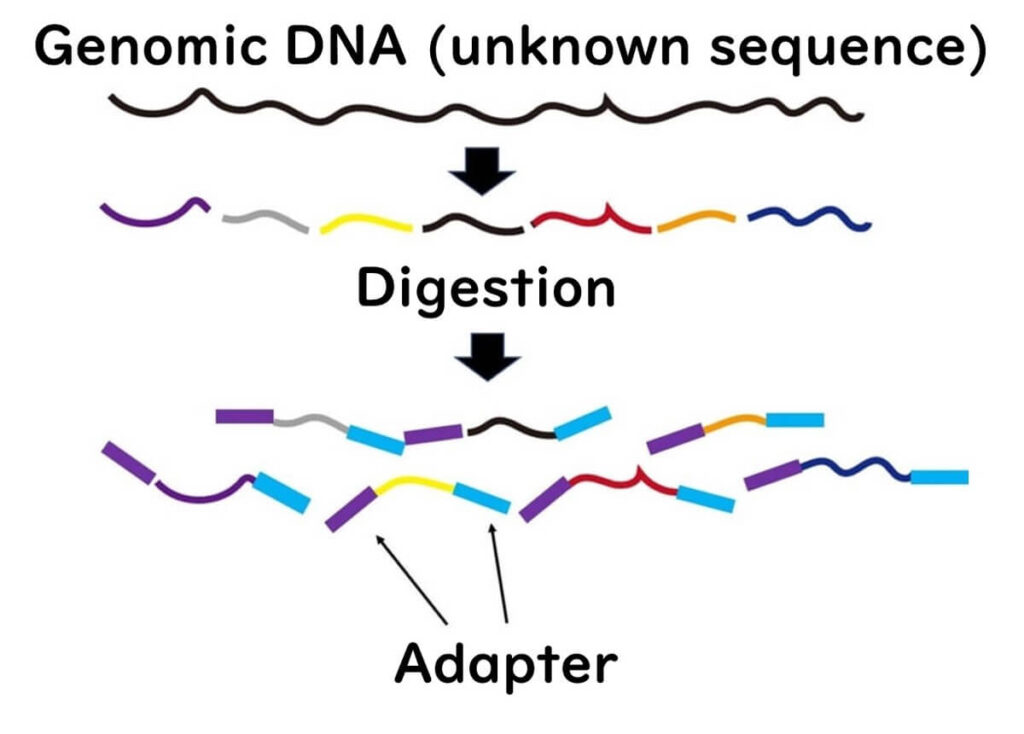
2️⃣ Adapter Ligation – Adapter sequences are attached to both ends of the fragmented DNA. This step is also used in Ion Torrent sequencing.
3️⃣ Flow Cell Attachment – The flow cell is pre-coated with DNA fragments complementary to the adapters, allowing the target DNA to bind.
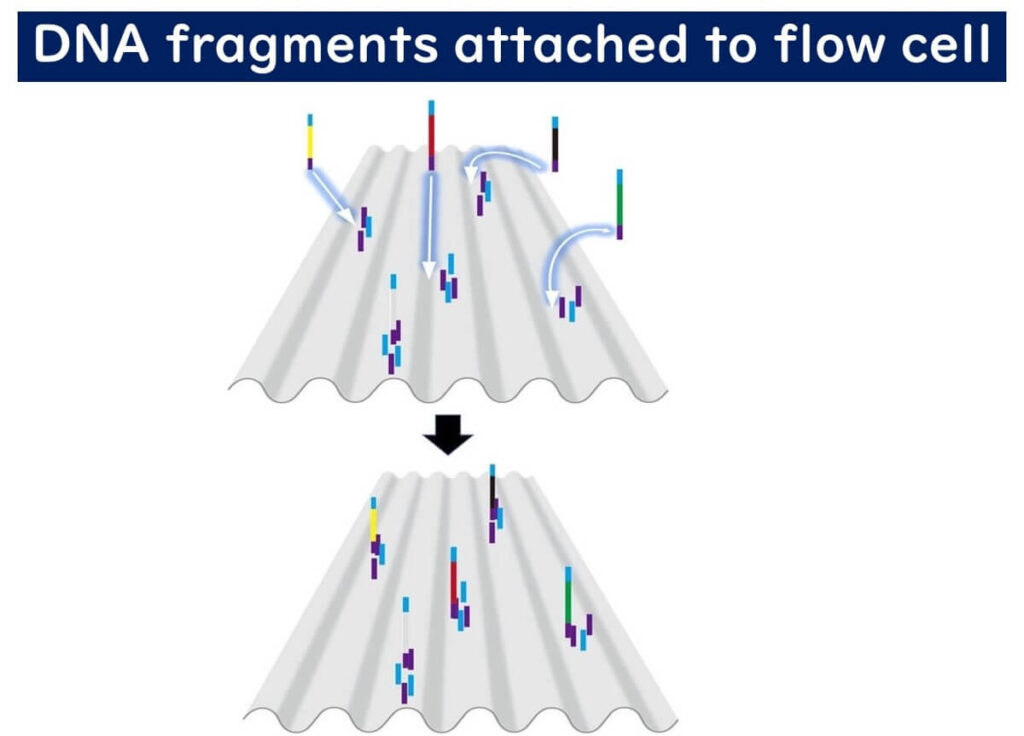
Bridge PCR: Amplification of DNA Clusters
Once bound to the flow cell, the DNA undergoes a unique amplification process called bridge PCR:
🔹 DNA extension – The bound target DNA acts as a template for complementary strand synthesis.
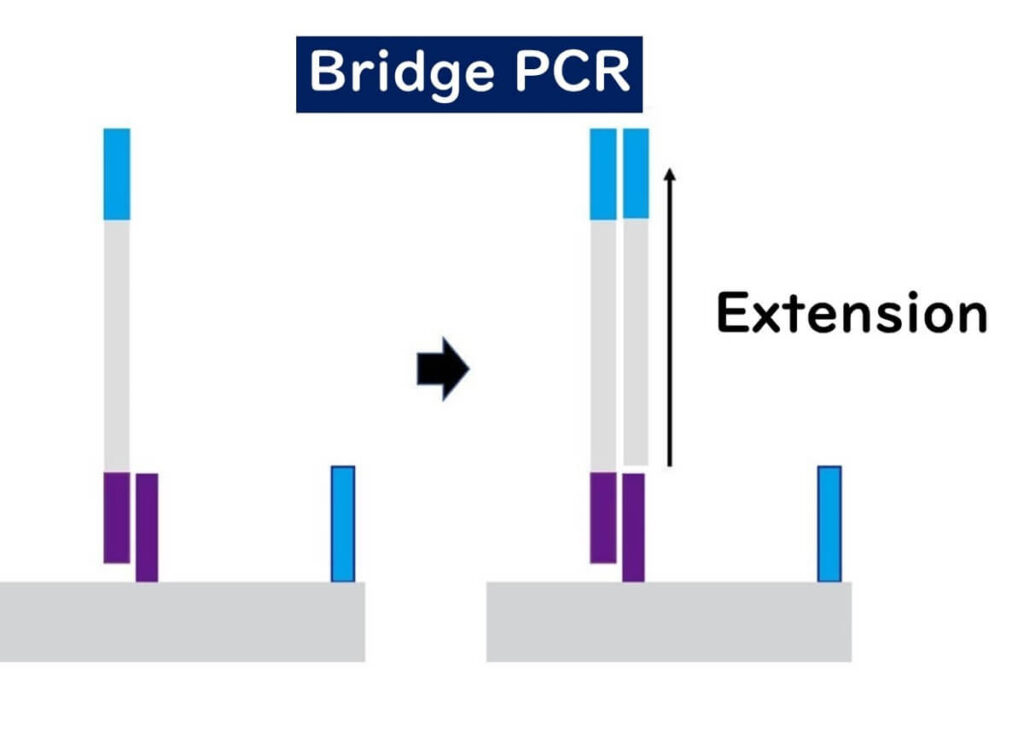
🔹 Bridge formation – The ends of the newly synthesized DNA fragments bind to nearby adapter sequences, creating bridge-like structures.
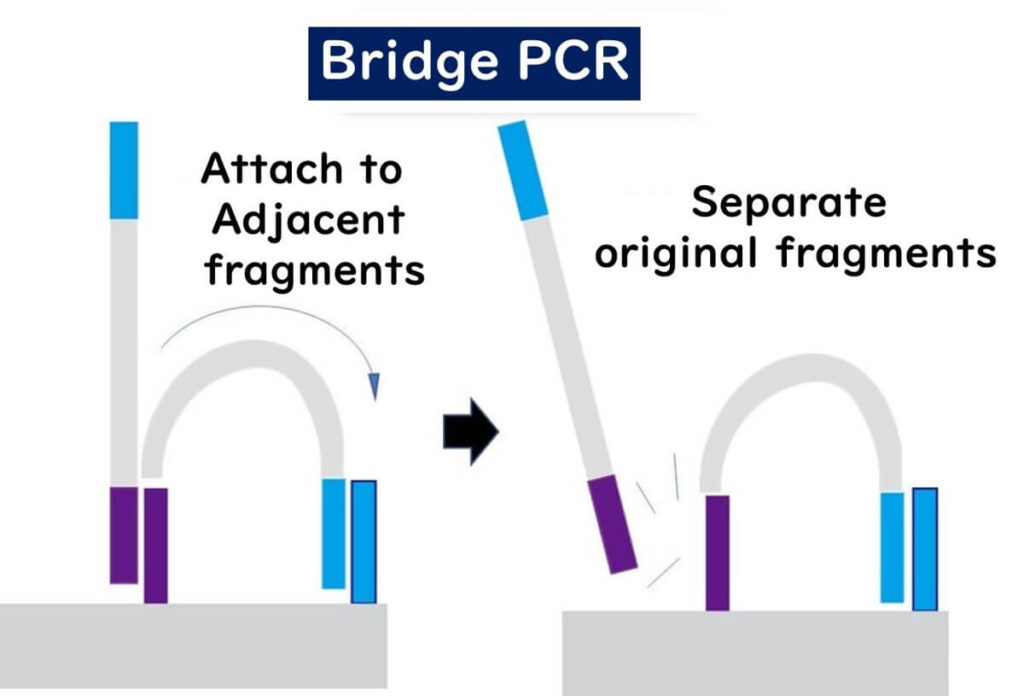
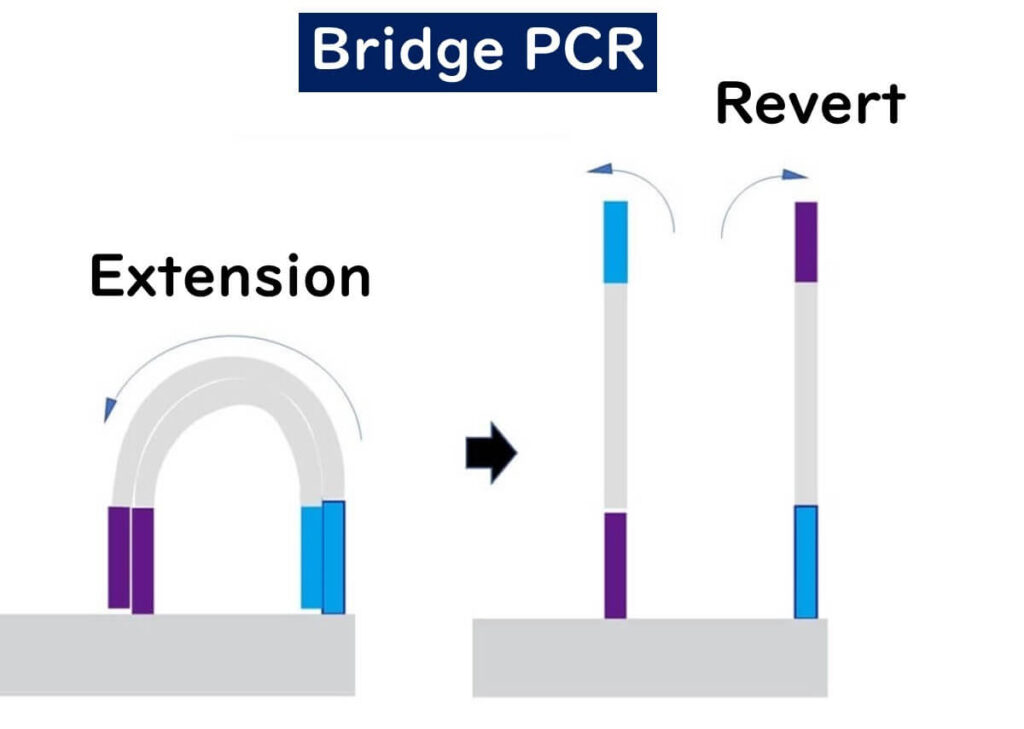
🔹 Repeated amplification – This cycle repeats multiple times, generating clusters of identical DNA fragments.
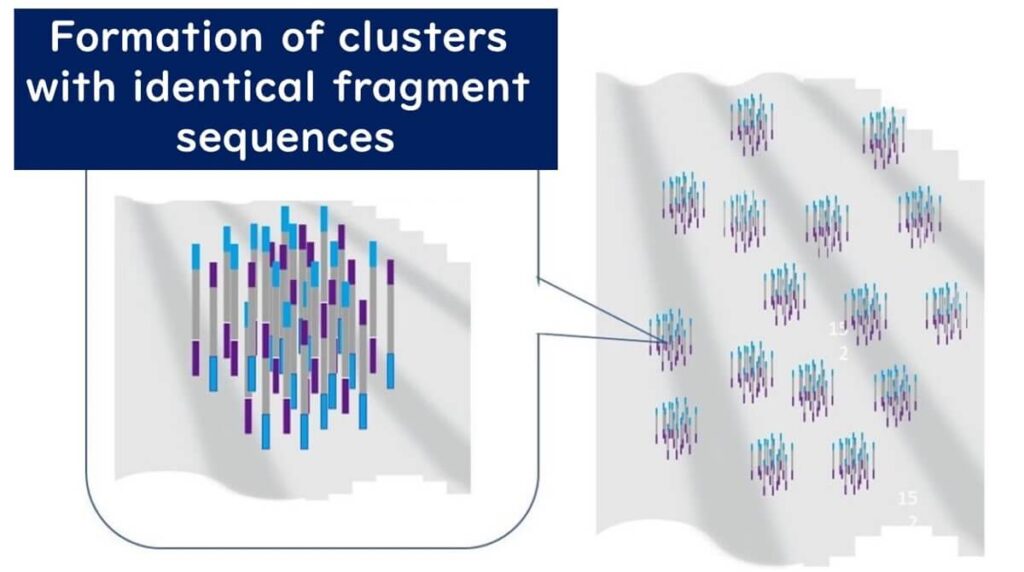
These clusters ensure high signal intensity, allowing millions of DNA sequences to be analyzed simultaneously.
How Illumina Sequencing Reads DNA
🔬 Fluorescent Labeling – Each nucleotide (A, T, C, G) is tagged with a unique fluorescent dye, similar to capillary sequencing.
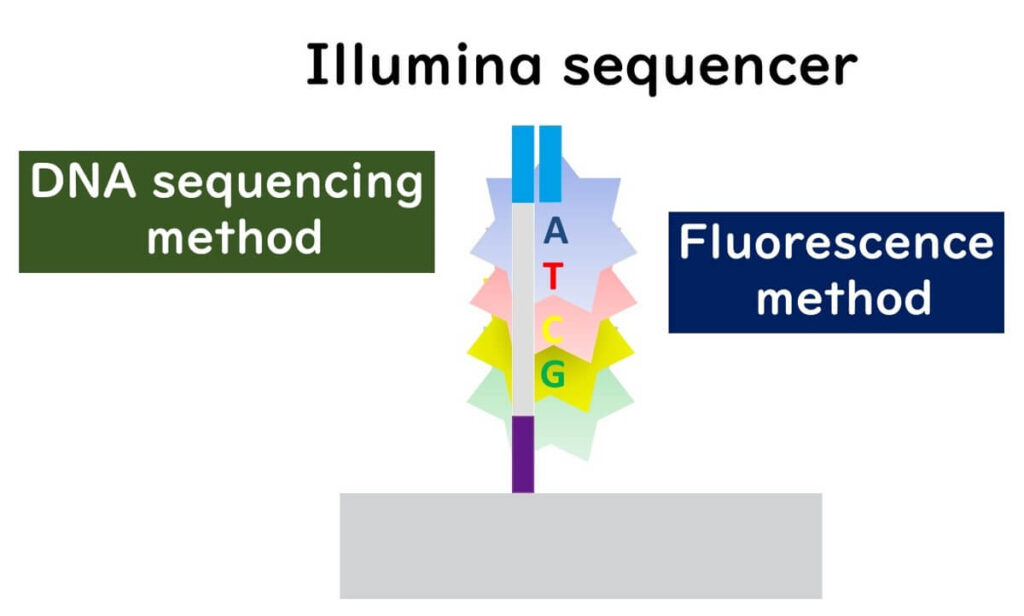
💡 Stepwise Base Addition – The sequencer adds one nucleotide at a time, detects fluorescence, then removes the fluorescent tag and blocking group before adding the next base.
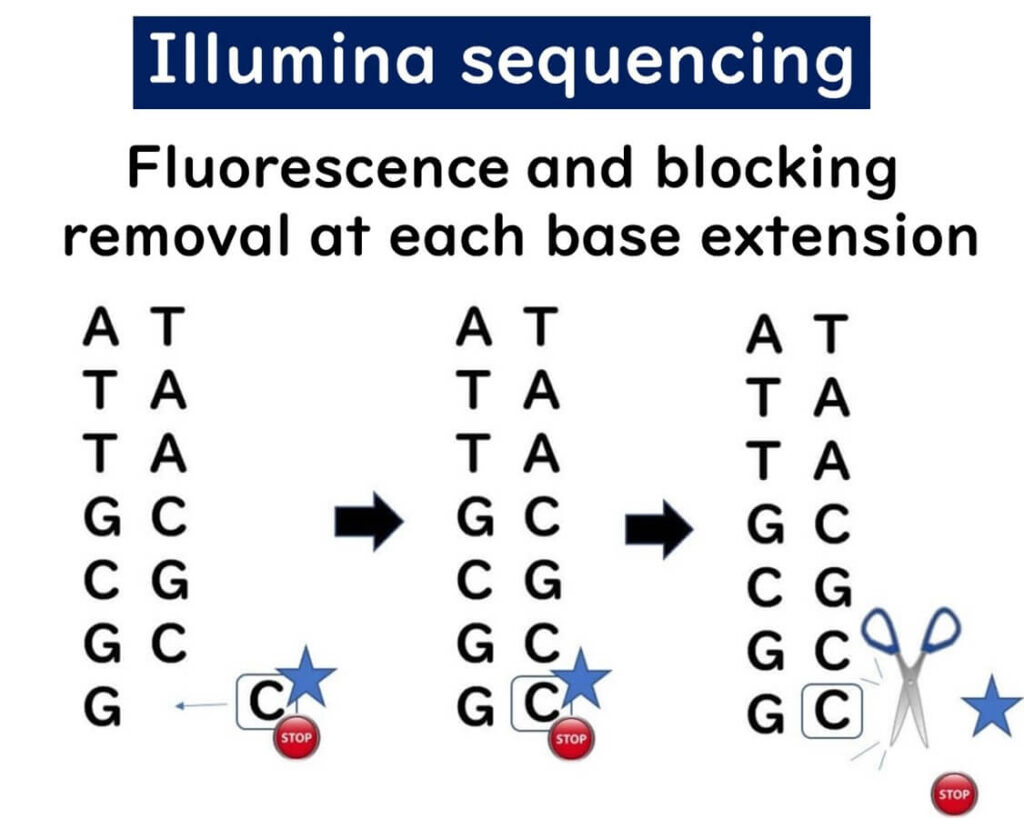
🎯 Error Reduction – This stepwise approach minimizes sequencing errors, making it ideal for SNP (single nucleotide polymorphism) analysis.

Advantages & Limitations of Illumina Sequencing
✅ High accuracy – Minimal sequencing errors due to stepwise base detection.
✅ Massive parallel sequencing – Millions of DNA fragments sequenced simultaneously.
⚠️ Limited read length – Reagents degrade over time, making long-read sequencing less efficient.

Watch the Illumina Sequencing Process
How Ion Torrent Sequencing Works: A Semiconductor-Based NGS Technology
Ion Torrent sequencing is a unique Next-Generation Sequencing (NGS) technology that differs from Illumina sequencing by using semiconductor-based detection instead of fluorescent dyes. This method allows for rapid and cost-effective DNA sequencing, making it a valuable tool in food microbiology and genetic research.
Key Steps in Ion Torrent Sequencing
1️⃣ DNA Fragmentation & Adapter Ligation
- The target DNA is fragmented using restriction enzymes.
- Adapter sequences are attached to both ends of the DNA fragments.
- This step is similar to Illumina sequencing.
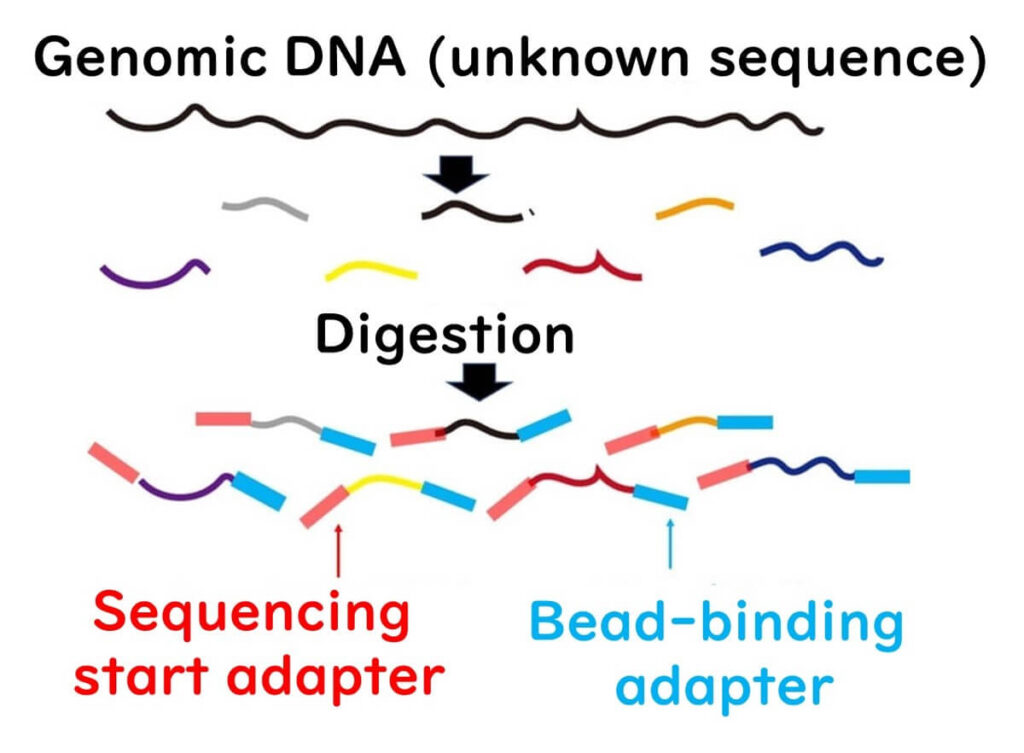
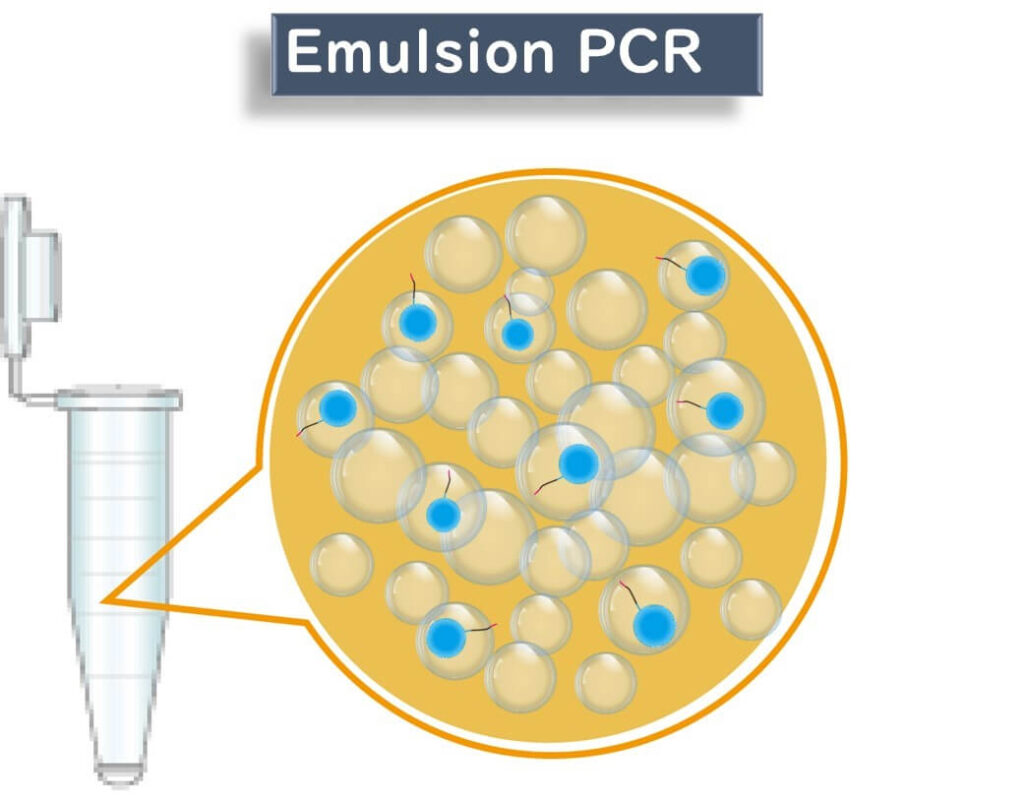
2️⃣ Emulsion PCR (ePCR) for DNA Amplification
- The fragmented DNA is bound to microbeads coated with complementary adapter sequences.
- The PCR reaction mixture is emulsified with oil, creating tiny water droplets, each acting as an individual PCR reactor.
- Each droplet ideally contains only one DNA molecule, ensuring precise sequencing.
- The DNA undergoes PCR amplification on the bead, creating multiple copies of the same sequence.
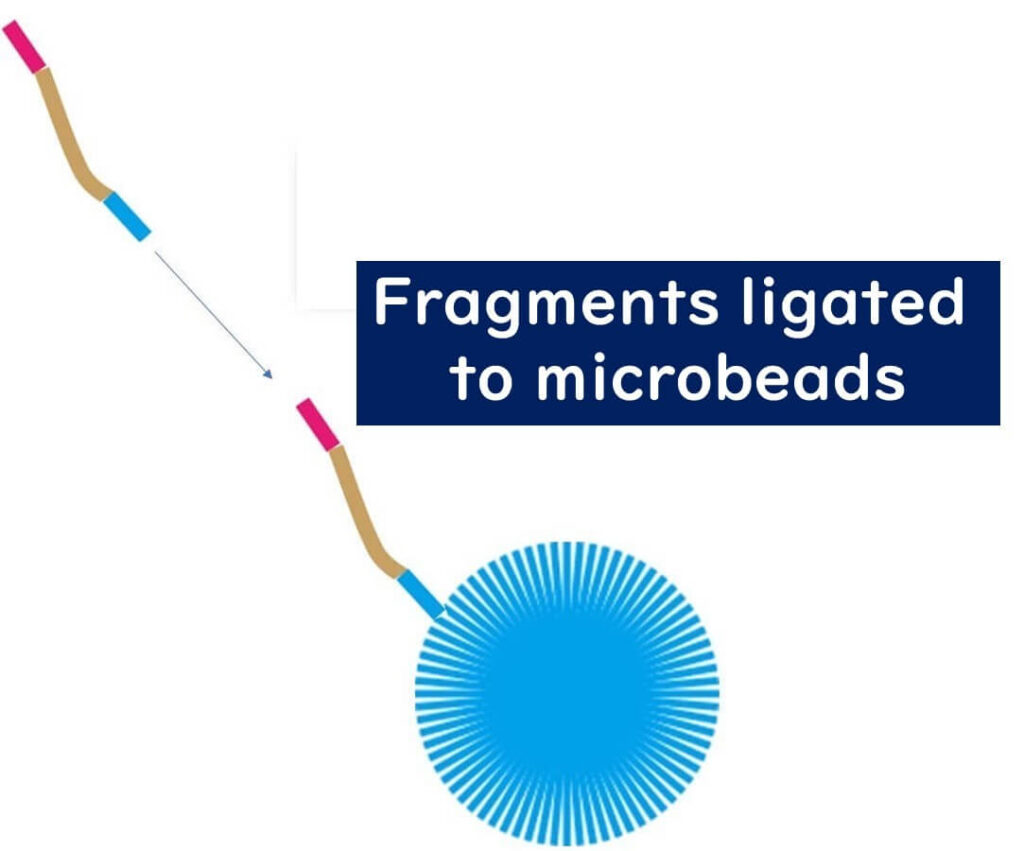
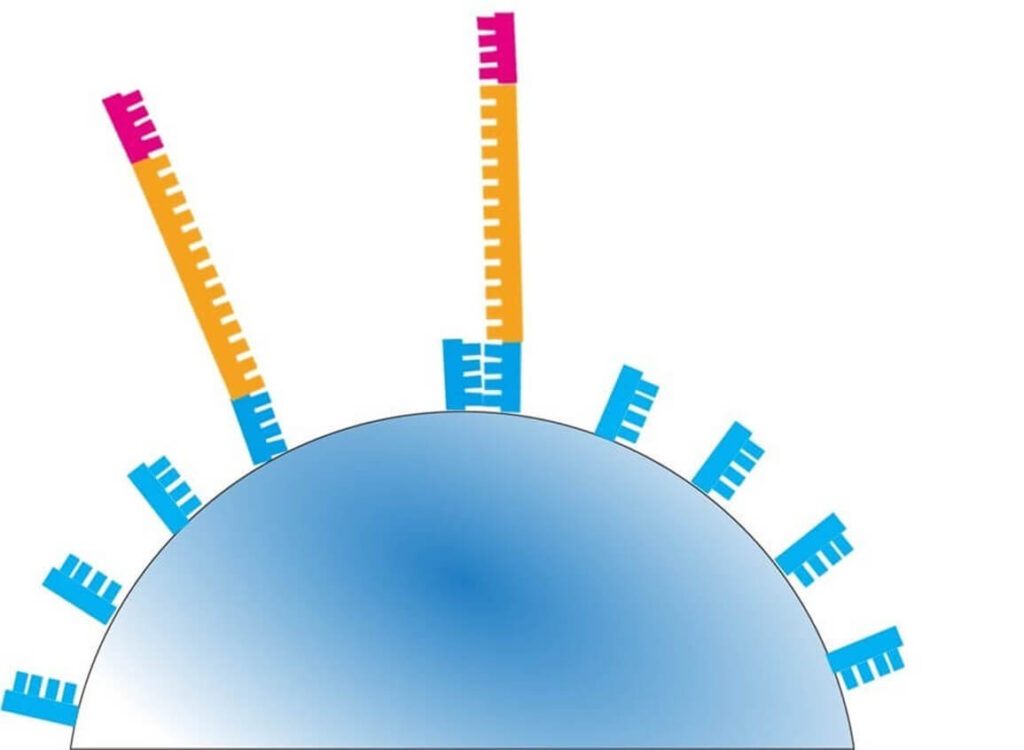
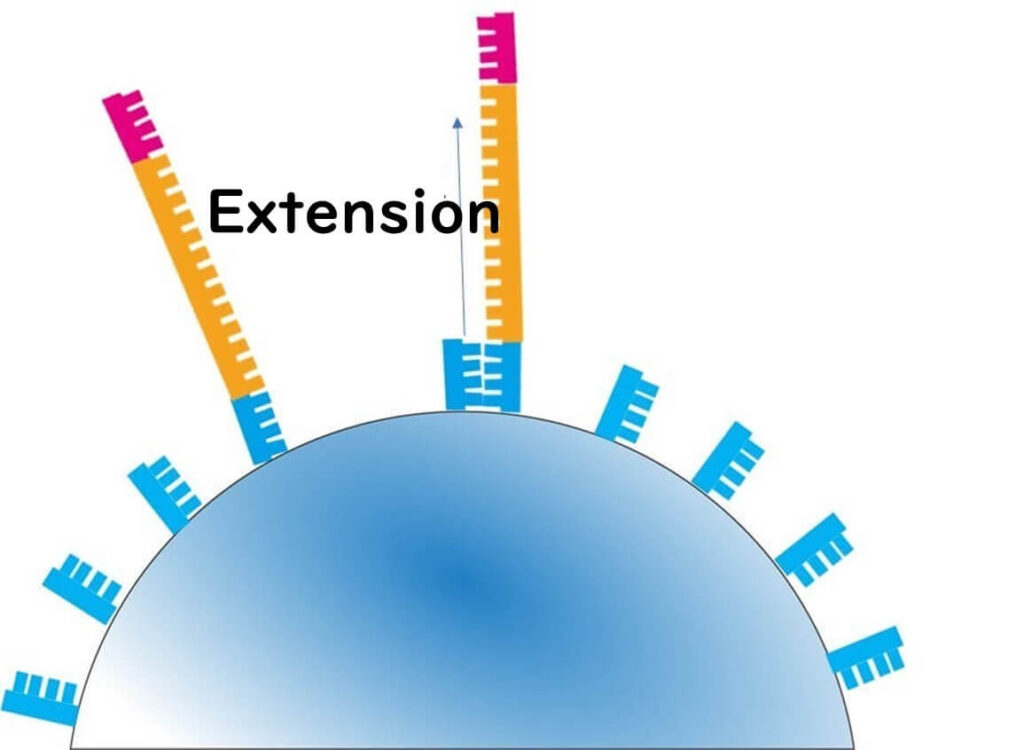
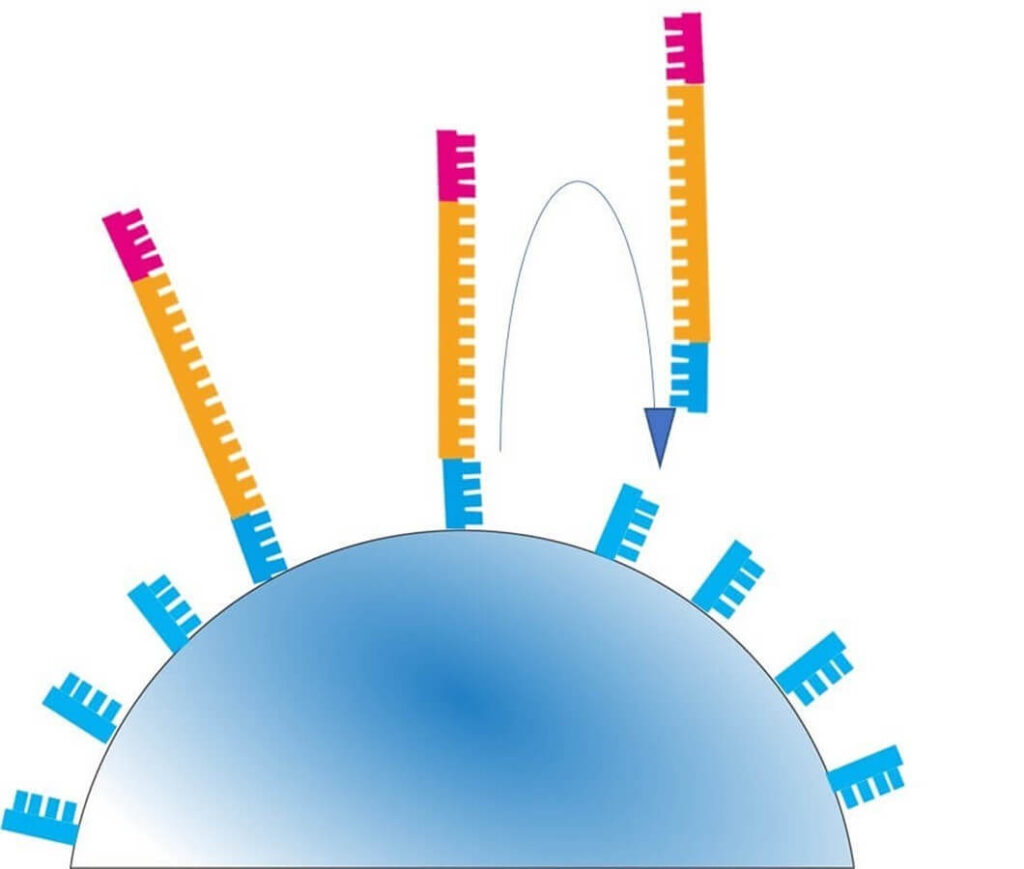
3️⃣ Bead Loading onto the Semiconductor Chip
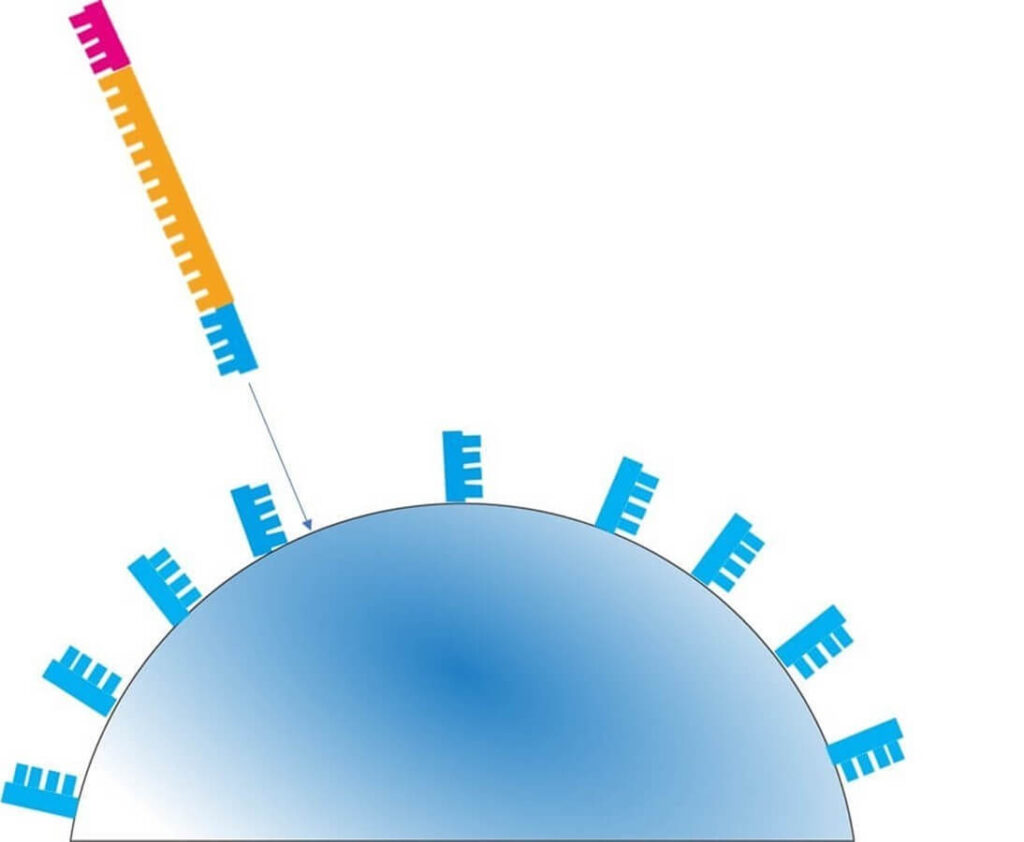
- Each amplified bead is placed into a well on a special semiconductor chip.
- The chip contains approximately 12 million wells, allowing for massively parallel sequencing.
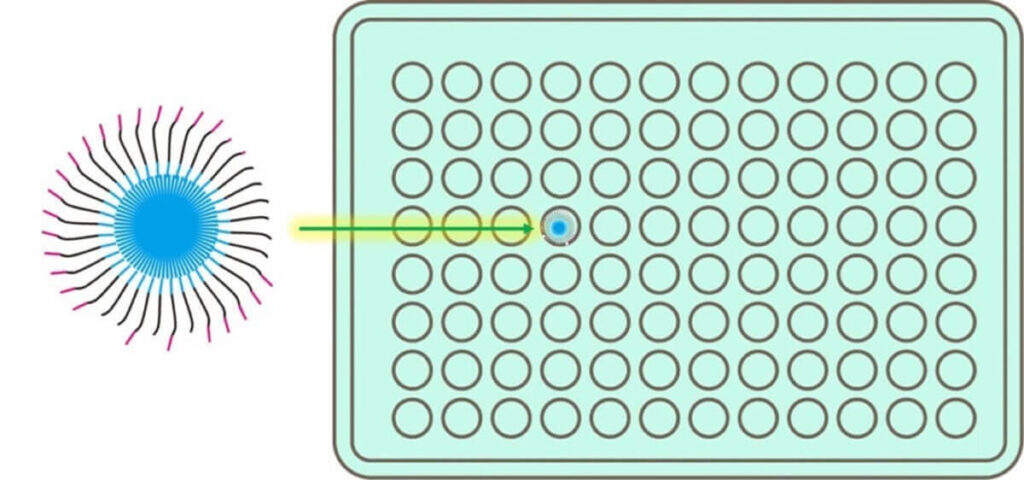
Ion Torrent Sequencing: A pH-Based Detection System
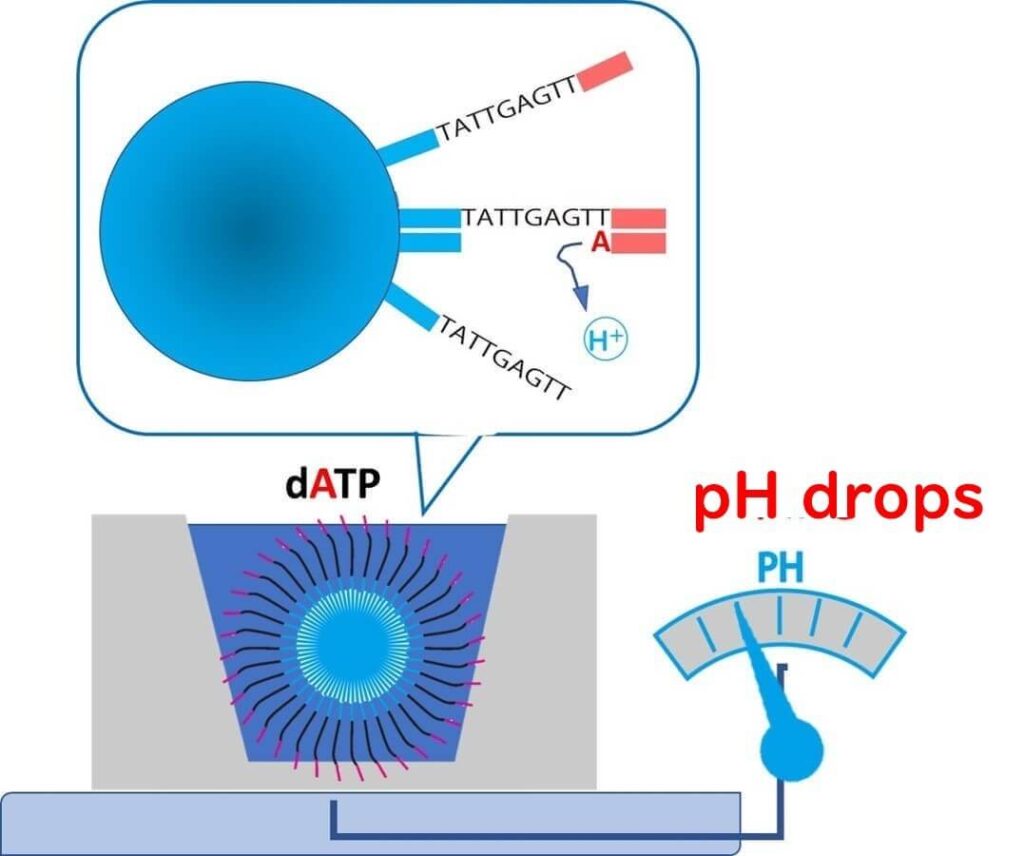
Unlike traditional sequencing technologies that rely on fluorescence detection, Ion Torrent sequencing determines DNA sequences by measuring pH changes caused by the release of H⁺ ions during nucleotide incorporation.
How It Works:
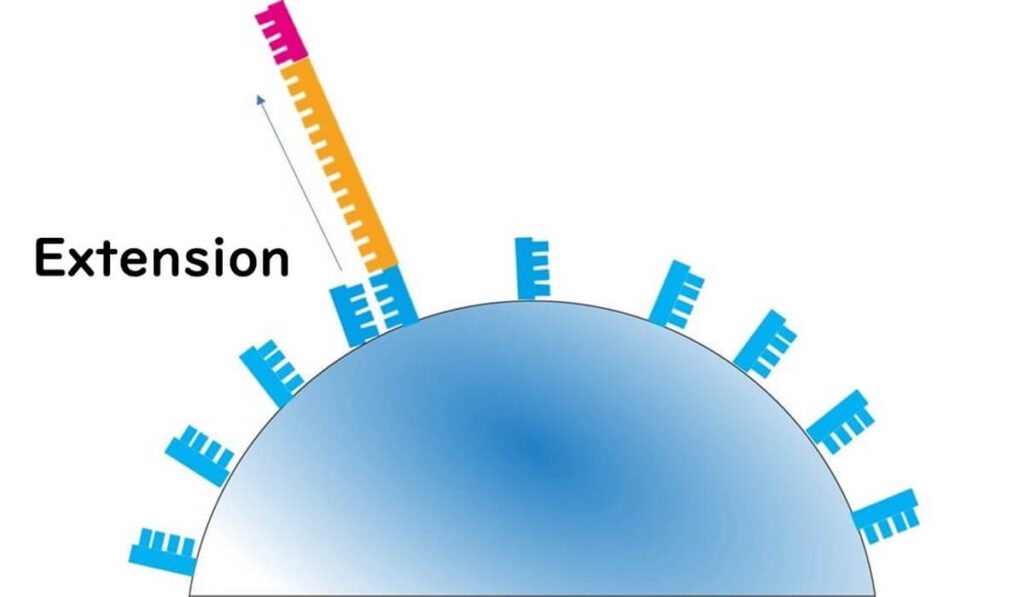
🔬 Step 1 – A reaction solution containing nucleotide A is added to the well.
- If the template DNA base is T, the polymerase extends the strand, releasing H⁺ ions.
- The pH decreases, and this change is detected with high sensitivity.
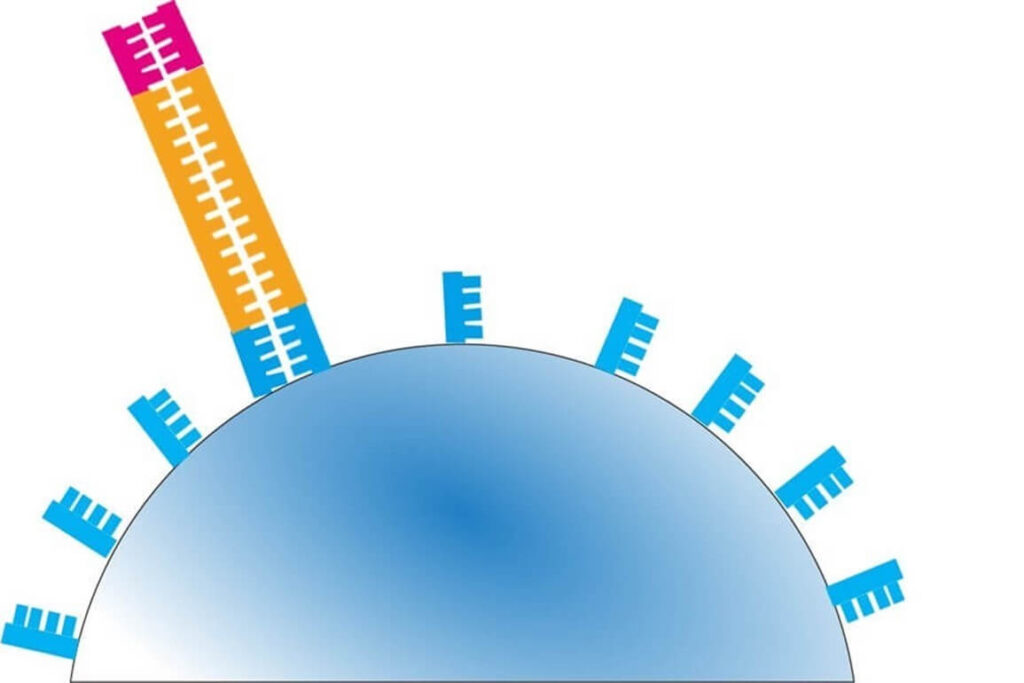
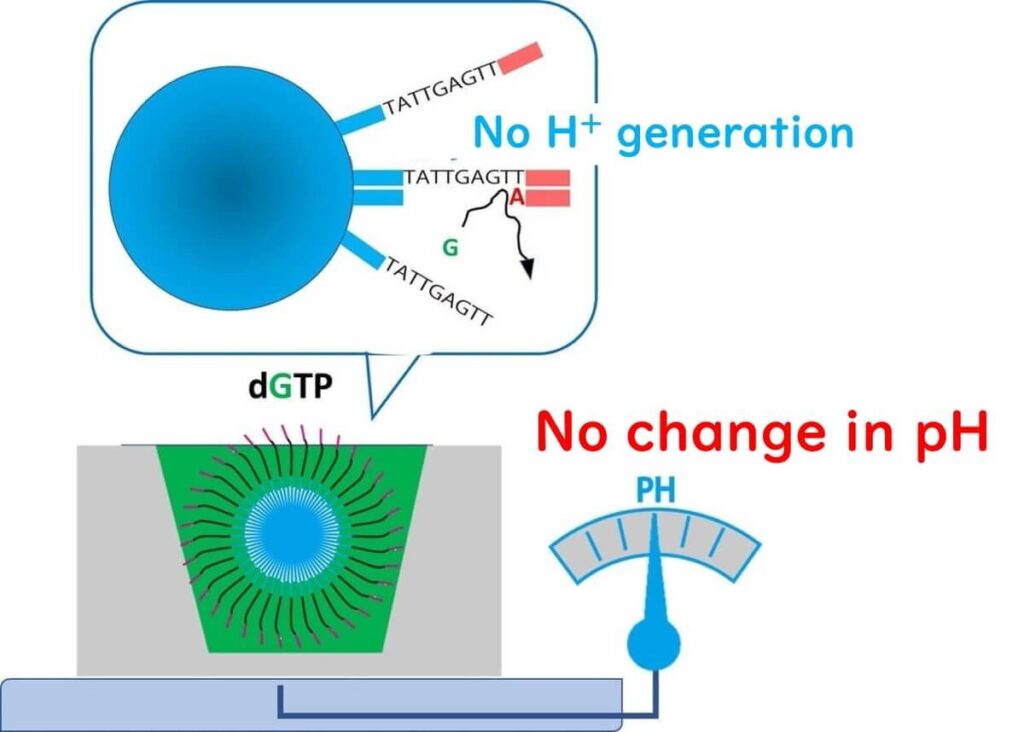
⚡ Step 2 – A reaction solution containing nucleotide G is added.
- If the next template base is not C (complementary to G), no extension occurs.
- As a result, no H⁺ ions are released, and the pH remains unchanged.
💡 Step 3 – This process is repeated across 12 million wells in parallel, generating massive amounts of sequence data.
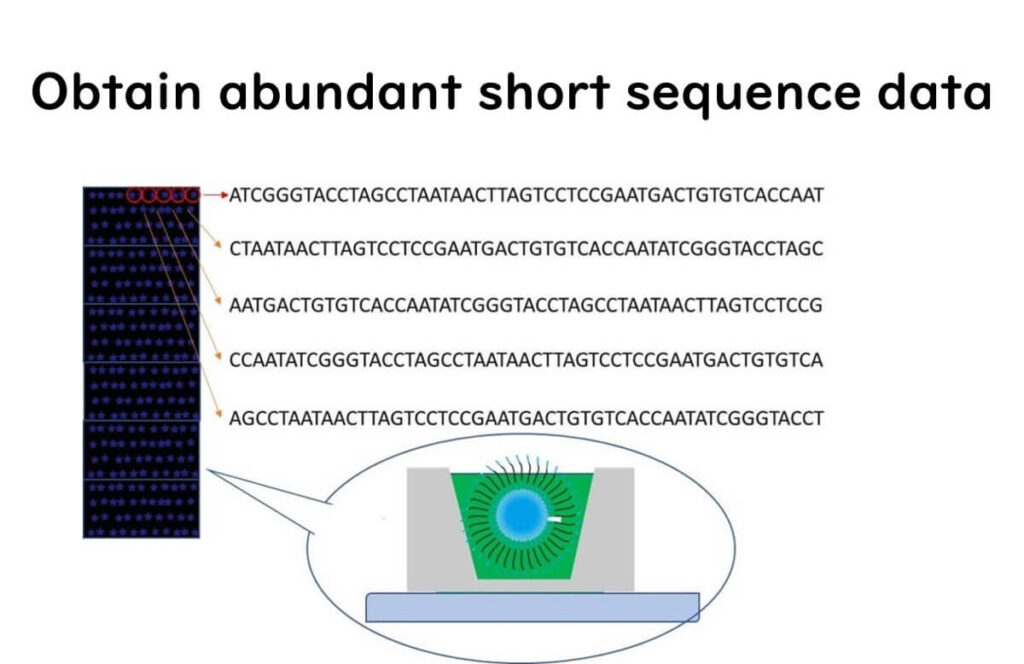
Once sequencing is complete, bioinformatics software reconstructs the full-length DNA sequence from these short fragments.
Advantages & Limitations of Ion Torrent Sequencing
✅ No fluorescence required – Uses semiconductor chips, reducing equipment costs.
✅ Fast sequencing – Directly detects pH changes, eliminating the need for optical scanning.
⚠️ Higher error rate – Homopolymer sequences (repeated bases like "AAAA") can be misread.

Watch the Ion Torrent Sequencing Process
Next-Generation Sequencing (NGS) in Food Microbiology Testing
Next-Generation Sequencing (NGS) has been transforming food microbiology testing for over 15 years. Compared to the Sanger method, NGS offers vastly higher sequencing speeds at significantly lower costs, making it a powerful tool for detecting and analyzing microorganisms in food.
In food microbiology, NGS technologies can be categorized into two major approaches:
1️⃣ Whole Genome Sequencing (WGS) of Bacterial Colonies
- The entire genome of a single bacterial isolate is sequenced.
- Used for strain identification, outbreak investigation, and antimicrobial resistance profiling.
2️⃣ Metagenomic Sequencing of Microbial Communities
- Comprehensive extraction of genomic data from multiple microorganisms in a food sample.
- Enables microbiome analysis, offering insights into food contamination sources and spoilage risks.
These approaches are revolutionizing food microbiology, providing unprecedented precision in microbial detection and characterization.
Advancements in Whole Genome Sequencing (WGS)
In the past decade, and especially in the last five years, significant technological advancements have improved genome sequencing from bacterial colonies. One major breakthrough is the development of core genome multilocus sequence typing (cgMLST).
Why is cgMLST Important?
✅ Standardized global database – Allows laboratories worldwide to share and compare genetic data.
✅ Enhanced outbreak tracking – Provides precise microbial strain identification, essential for foodborne disease surveillance.
✅ Potential for routine food industry testing – While initially developed for public health institutions, cgMLST is increasingly applicable to food safety monitoring in private companies.
Metagenomics and 16S rRNA Sequencing in Food Microbiology
Unlike WGS, which focuses on a single bacterial isolate, metagenomic sequencing analyzes the entire microbial community within a food sample. This technique, particularly through 16S rRNA amplicon sequencing, has revolutionized:
🔬 Food Quality Control – Identifies microbial profiles in processed foods and raw ingredients.
⚡ Rapid Contamination Detection – Enables fast identification of spoilage organisms without traditional culture methods.
🌍 Microbiome Research – Provides insights into bacterial ecosystems in food production environments.
The speed and depth of analysis provided by NGS far surpass traditional microbiological culture methods, making it an essential tool for modern food safety and quality management.
potential for future applications, especially in fields such as food quality control and raw material management.
Third-Generation Sequencing (TGS): Nanopore Technology in Food Microbiology
To overcome the limitations of Next-Generation Sequencing (NGS)—specifically its short read lengths (~300 bp)—Third-Generation Sequencing (TGS) was developed. TGS enables long-read sequencing (over 100 Kbp), making it a game-changer in genomic research and food microbiology testing.
Since around 2017, third-generation sequencing technologies have rapidly expanded worldwide, with two leading platforms:
🔹 Pacific Biosciences (PacBio) – Uses single-molecule real-time (SMRT) sequencing.
🔹 Oxford Nanopore Technologies (ONT) – Uses nanopore-based sequencing, offering real-time and portable sequencing capabilities.
Unlike Sanger sequencing and NGS, nanopore sequencing does not require nucleotide synthesis, significantly reducing operational complexity and costs.
How Nanopore Sequencing Works
In nanopore sequencing, DNA or RNA molecules pass through a nanopore, a protein embedded in a polymer membrane. This nanometer-scale pore allows ions to flow through when a voltage is applied.
As DNA moves through the nanopore, each nucleotide (A, T, C, G) disrupts the ion current in a unique way. These current changes are detected and translated into DNA sequences.
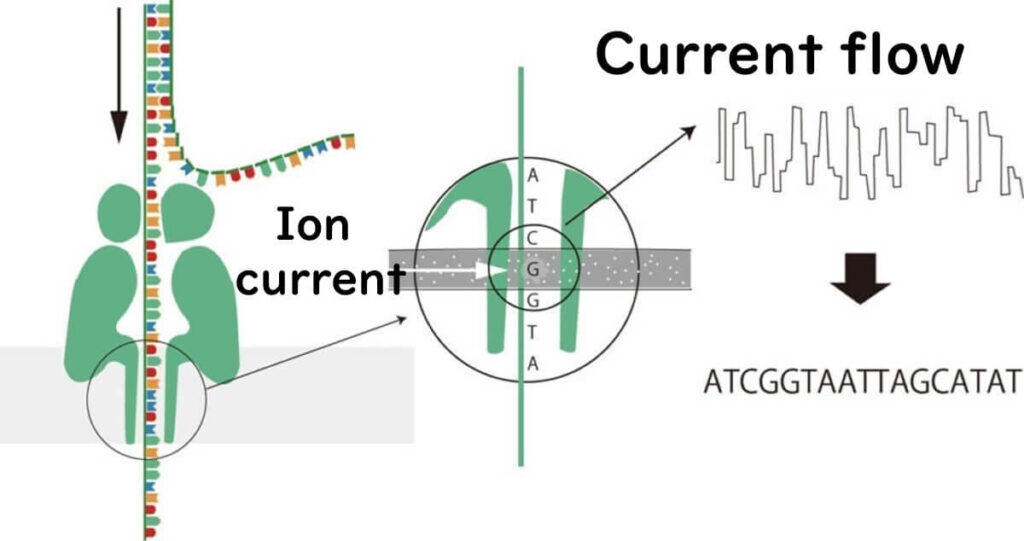
Key Differences from Other Sequencing Methods:
✅ No nucleotide synthesis – Unlike Sanger sequencing and NGS, no fluorescent dyes or chemical modifications are required.
✅ Real-time sequencing – DNA is read as it passes through the nanopore, allowing for instant data generation.
✅ Portable sequencing – Devices like MinION enable sequencing outside the lab, making them ideal for on-site food safety testing.
📺 For a detailed explanation, visit the Oxford Nanopore Technologies website.
Advantages of Nanopore Sequencing in Food Microbiology
1️⃣ Long-Read Sequencing
- Can read 10,000–50,000 bp or more in a single pass.
- Overcomes short-read limitations of NGS, allowing more accurate genome reconstruction.
- Enables species-level identification of microbial communities through full-length 16S rRNA sequencing.
2️⃣ No Nucleotide Synthesis (Simplified Sample Preparation)
- Directly sequences non-amplified genomic DNA, eliminating the need for PCR, enzymes, or cloning.
- Lower running costs & faster turnaround compared to NGS.
- Portable devices like MinION enable real-time sequencing for pathogen detection in food factories.
3️⃣ On-Site Microbial Testing
- Ideal for rapid detection of Listeria and Salmonella contamination.
- Can be applied to quality control and foodborne outbreak investigations.
Challenges of Nanopore Sequencing in Food Microbiology
⚠️ Higher Error Rates – Current nanopores lack sufficient resolution to distinguish individual nucleotides perfectly.
⚠️ Homopolymer Issues – Repeated sequences (e.g., "AAAA") are difficult to read accurately.
⚠️ Speed Control Needed – DNA movement through the nanopore must be slowed to millisecond precision for optimal accuracy.
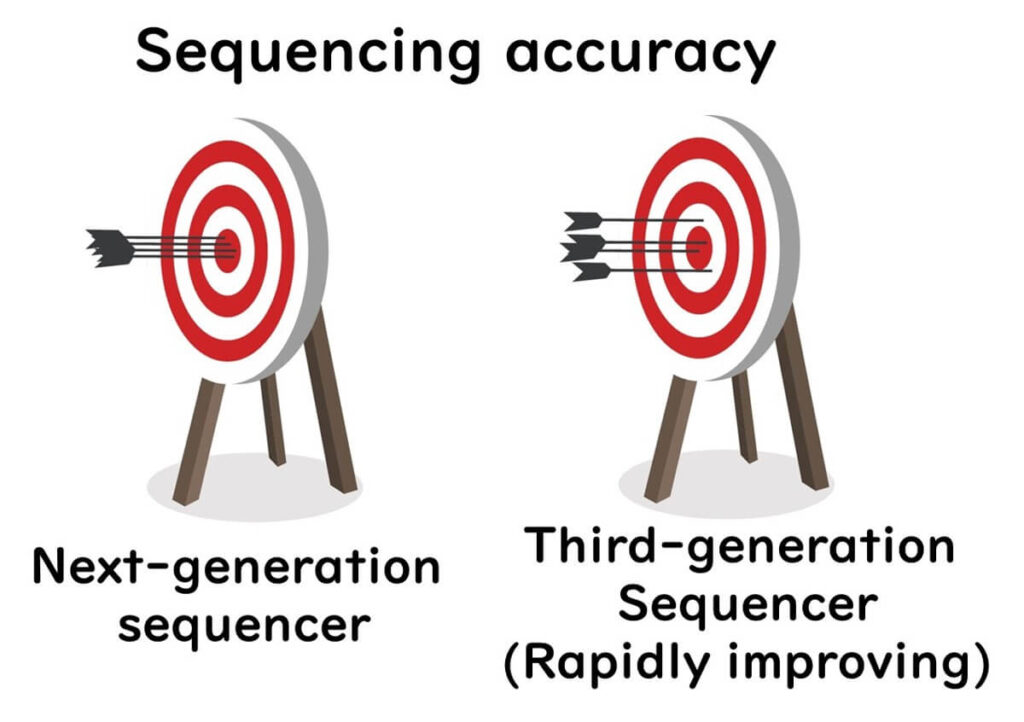
🔬 However, ongoing advancements in nanopore technology are addressing these limitations.
✅ New nanopores and improved algorithms have pushed sequencing accuracy close to 99.99%, making it increasingly viable for food microbiology applications.