PCR detection is a powerful tool in food microbiology, offering precision in microbial DNA detection. However, it also presents limitations that can impact accuracy and reliability. This page delves into critical challenges, including the detection of dead bacterial DNA, blind spots in gene detection, and the risks of false positives and negatives. By recognizing these limitations, food safety professionals can make informed decisions and implement strategies to enhance testing reliability.
Introduction: Understanding the Limitations of PCR Testing in Food Microbiology
PCR (Polymerase Chain Reaction) is an indispensable tool in food microbiology, known for its precision in detecting microbial DNA. However, like any scientific method, PCR has its limitations and challenges. This article delves into some of the most critical aspects of PCR testing, including its blind spots, practical issues, and key considerations for accurate microbial detection. By understanding these limitations, food safety professionals can better interpret results and make informed decisions in quality control and assurance.
Recognizing Only the Ends of Genes
It's crucial to understand that PCR detection only targets and recognizes the sequences at the ends of the target gene.
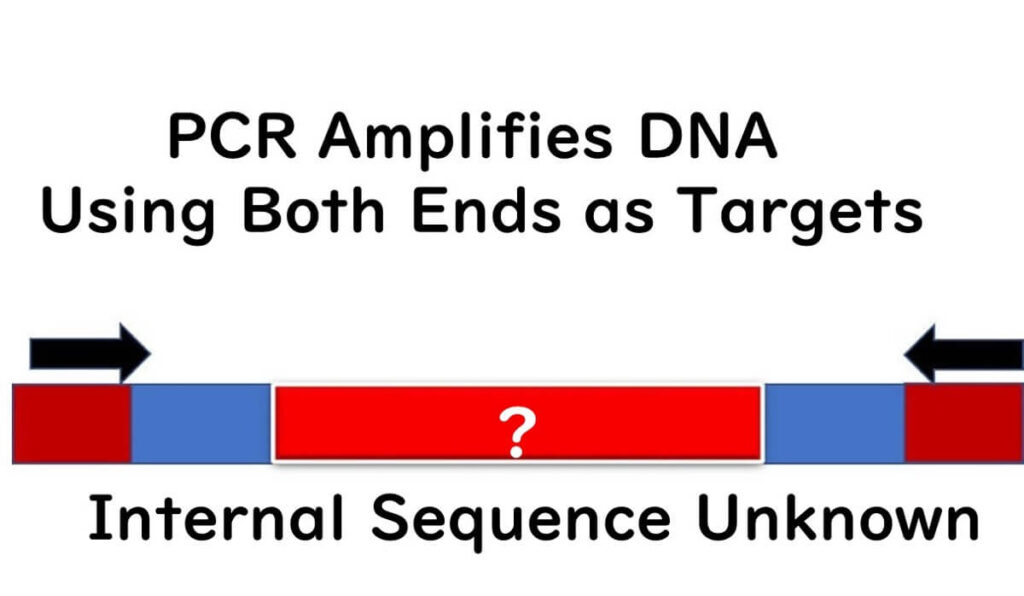
When conducting PCR tests targeting specific pathogenic genes, even if the target gene is amplified, it does not necessarily mean that the gene is expressed in the cells or functioning as a pathogen.
This highlights a critical limitation of PCR testing—it is not always based on an infallible logical foundation for detection. For food safety professionals, this distinction is vital when interpreting PCR results.
No Viable Bacteria Detected: Challenges in Differentiating Live and Dead Cells
DNA Remains Intact Even After Microbial Cell Death
One of the major limitations of genetic testing methods is the inability to differentiate between DNA from live and dead cells. When DNA is boiled at 100°C, the hydrogen bonds holding the double strands together break, resulting in single strands. However, the phosphodiester bonds linking the deoxyribose sugars remain intact.
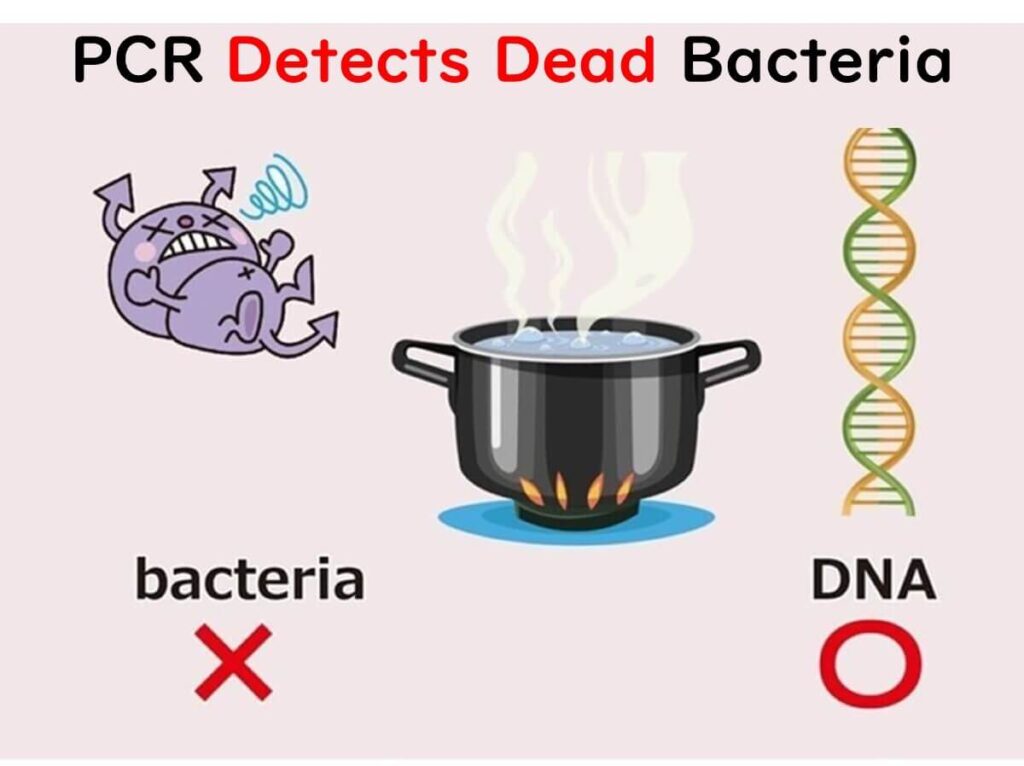
This means that even if microorganisms are killed by heat, their DNA sequences remain detectable. As a result, PCR tests used to directly examine microorganisms in food cannot determine whether the microorganisms are viable or non-viable.
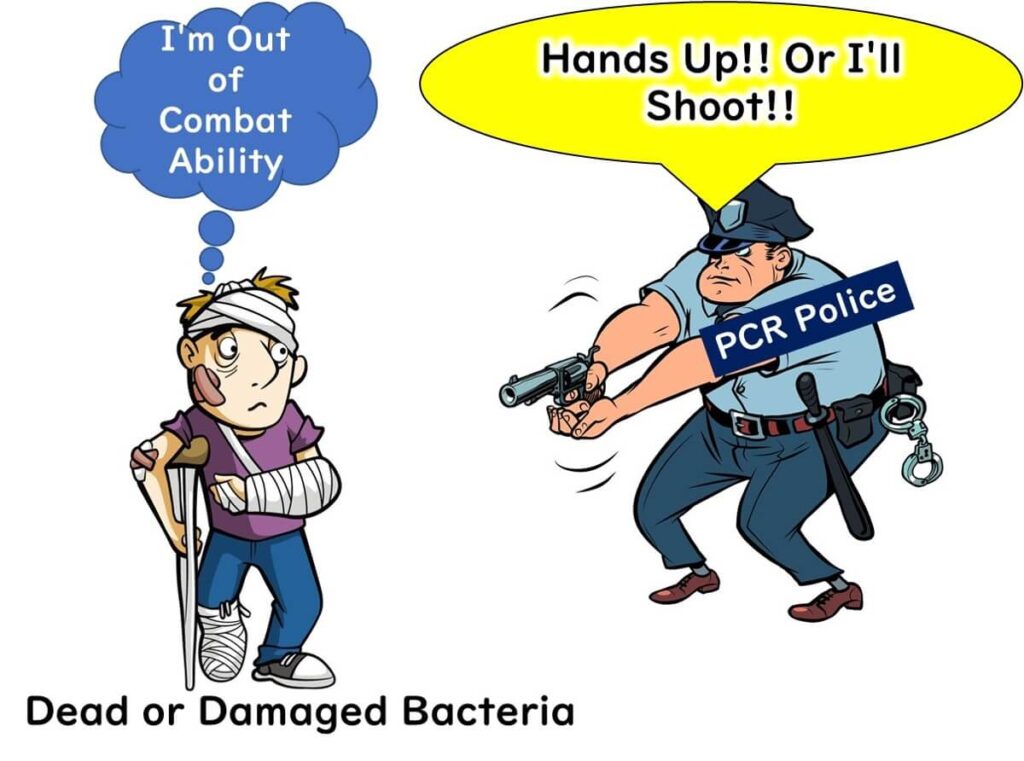
Recent research has investigated the use of nucleic acid intercalating dyes, such as Ethidium Monoazide (EMA) and Propidium Monoazide (PMA), to address this issue. These dyes selectively penetrate dead bacterial cell membranes and bind to DNA, inhibiting PCR amplification. Consequently, only DNA from live bacteria is amplified. However, the effectiveness of these methods depends on the extent of cell damage, and practical implementation is often limited by legal and regulatory considerations.
Overcoming the Live Bacteria Detection Issue with Enrichment Cultures
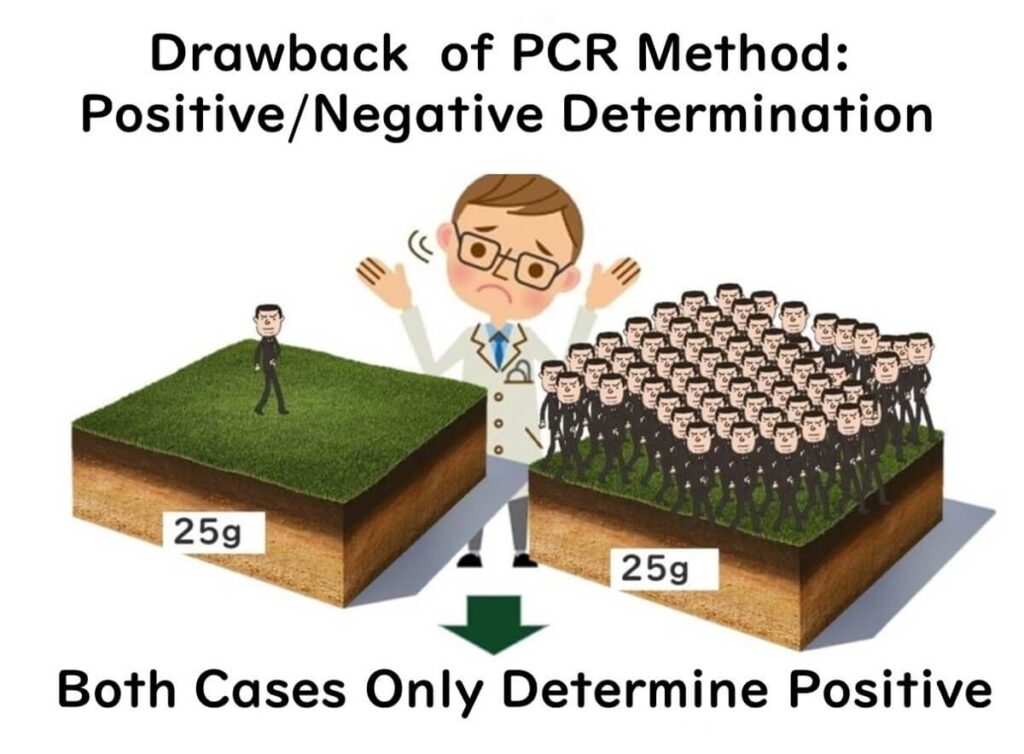
In practical food testing, the limitations of genetic testing are often mitigated through enrichment cultures. PCR testing alone is typically not sensitive enough to detect foodborne pathogens at levels as low as one cell per 25 grams of food. Enrichment cultures, which increase the number of target bacteria to detectable levels, are thus essential in most cases. These cultures provide reliable results for determining the presence of live bacteria.
Note: For more details on why PCR methods cannot reliably detect pathogens at the level of one cell per 25 grams of food, refer to the article on "Real-Time PCR in Food Microbiology: Challenges and Applications"
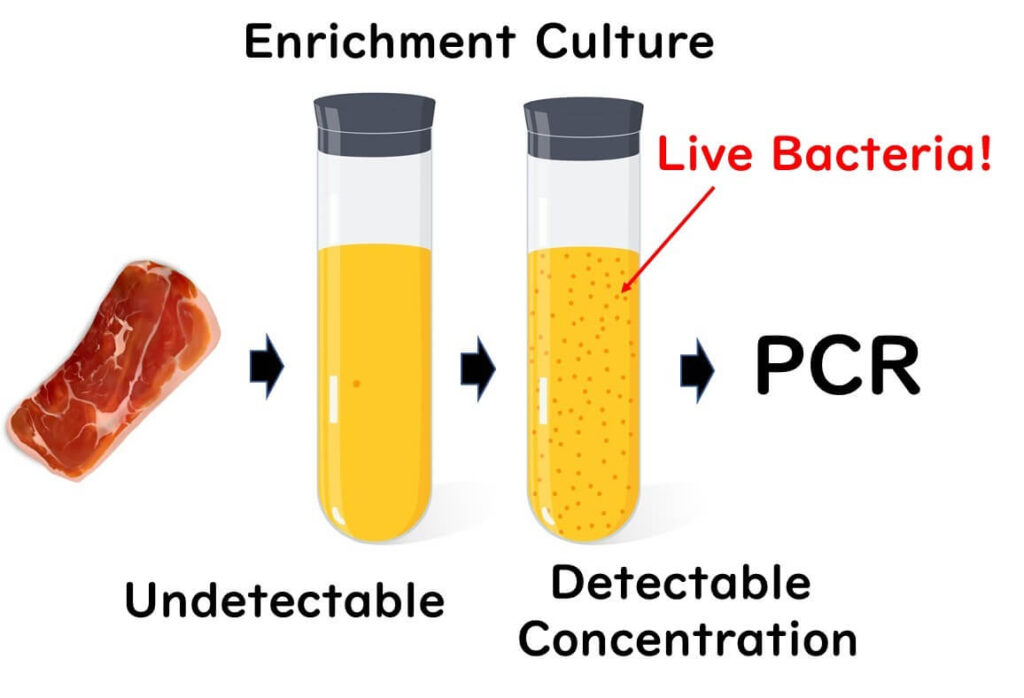
While enrichment cultures are effective in confirming the presence of live bacteria, they do not provide quantitative information about the initial bacterial load—whether it was a single bacterium or several hundred in 25 grams of food.
The Blind Spots of Gene Detection: Understanding False Positives in PCR
Gene Detection Challenges in PCR Testing
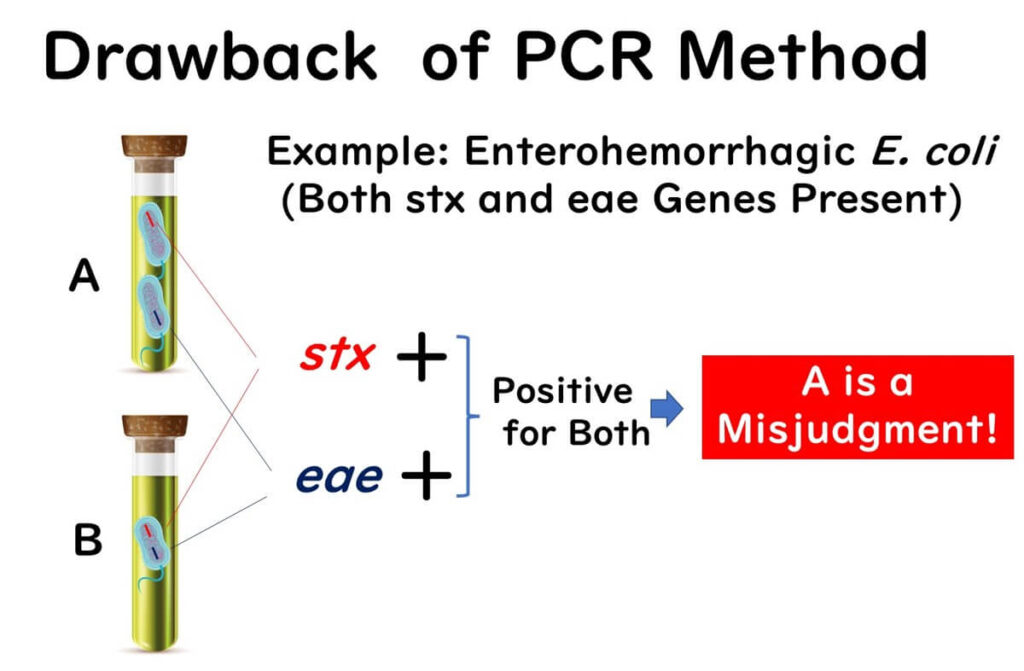
PCR testing is capable of detecting genes directly from enrichment cultures without the need to isolate colonies. However, this method has a significant blind spot, particularly in cases involving pathogenic Escherichia coli (E. coli). Detection often relies on the presence of two target genes, such as stx (Shiga toxin) and eae (intimin).
In some instances, an enrichment culture may contain separate E. coli strains, each carrying only one of these genes. This can result in a false positive, as the PCR test detects the presence of both genes without confirming they belong to the same bacterial strain. Traditional culture methods may subsequently fail to identify the pathogenic E. coli.
Observed Issues in Standards
This challenge is particularly notable in international ISO methods and domestic testing standards for enterohemorrhagic E. coli. Although PCR testing may yield positive results for these target genes, follow-up cultures often reveal negative outcomes. Such discrepancies underscore the blind spot inherent in gene detection methods.